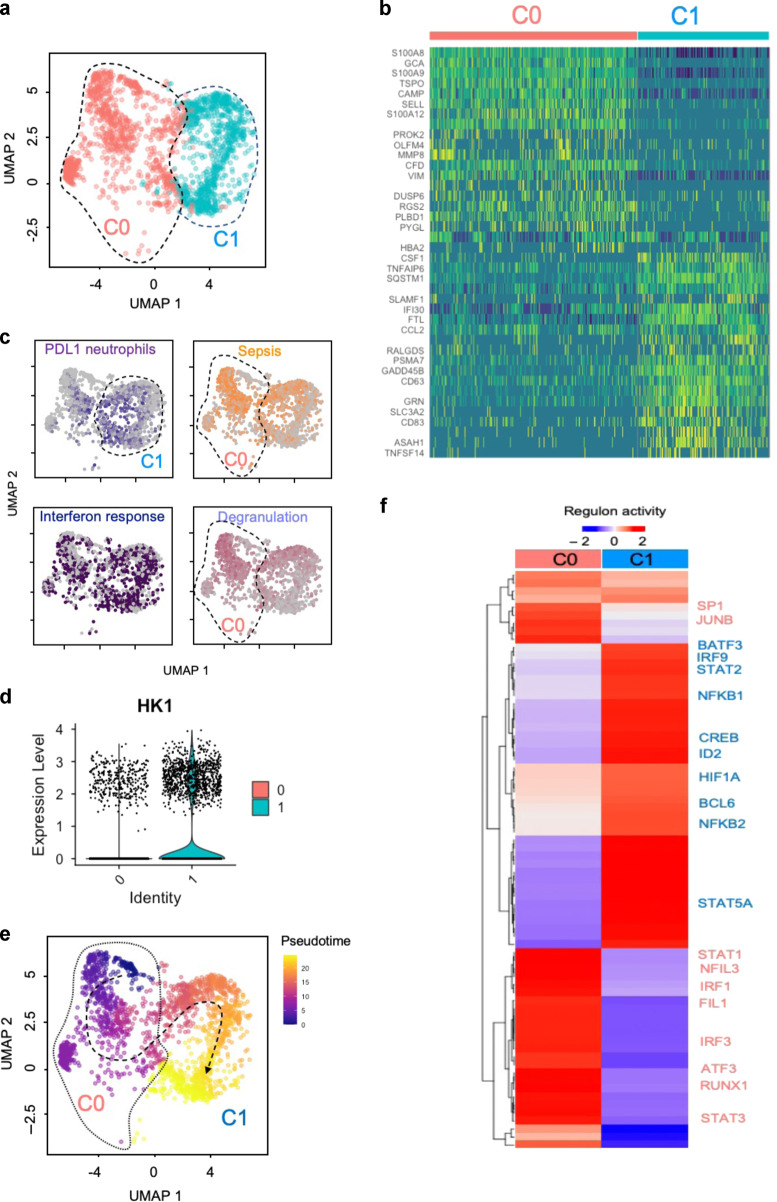
Fig. 4. Transcriptomic program of the neutrophils recruited in IA LPS-exposed lungs.
a UMAP embedding of neutrophils extracted from a larger dataset of lung immune cells colored by clusters. b Row-scaled expression of the highest DEGs in each cluster (Bonferroni-adjusted p values < 0.05). c UMAP embedding of neutrophils colored by average expression of genes associated with exhaustion (PD-L1, purple), sepsis (orange), interferon response (blue) and neutrophil degranulation (lavender). These gene signatures were derived from public gene expression datasets (see Methods). d Violin plot of HK1 expression in fetal lung neutrophils following IA LPS. e UMAP embedding of neutrophils colored by pseudotime with overlaid trajectory. f Row-scaled regulons activity for neutrophil clusters. k-means clustering was used to arrange clusters and regulons.