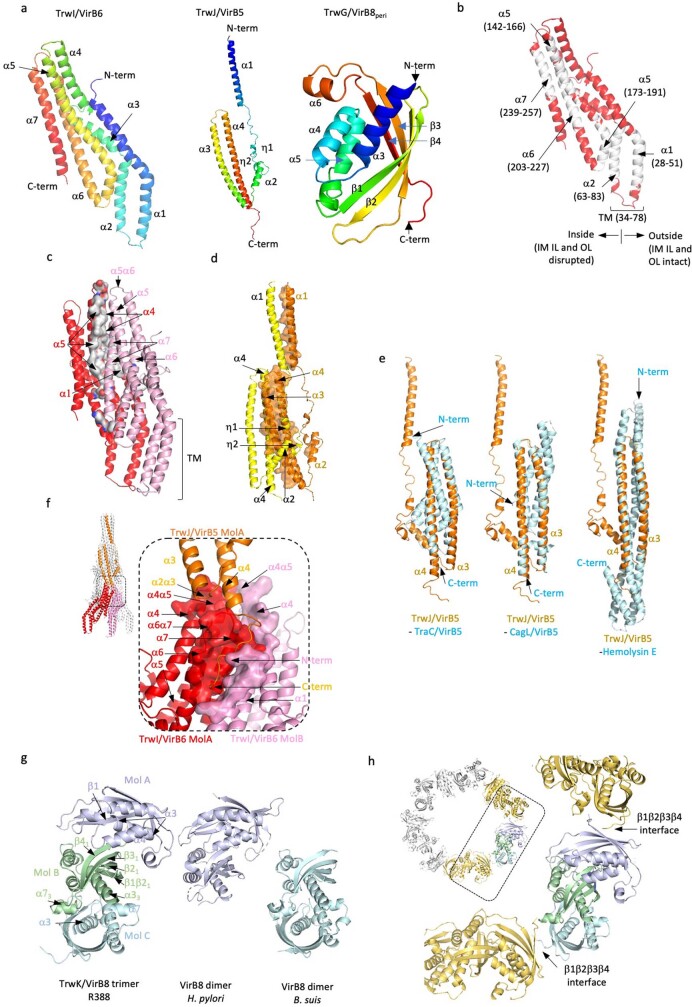
Extended Data Fig. 7. Details of Stalk and Arches proteins.
a, Secondary structure definition of TrwI/VirB6 (left), TrwJ/VirB5 (middle) and TrwG/VirB8peri (right). The ribbon for each protein is coloured in rainbow colours from dark blue for the N-terminus to red for the C-terminus. All secondary structures are labelled. b, Locations of the predicted hydrophobic TMs in TrwI/VirB6. The TMpred server58 suggests a number of potential TMs in TrwI/VirB6: these predicted TMs are here mapped in white onto the TrwI/VirB6 structure. Of these, only two are observed inserting in the inner membrane, α1 and α2. All α-helices that contain a hydrophobic region are labelled. Boundary residues for these regions and location of the TM region are indicated. Also, we observe that the region of the IM within the TrwI/VirB6 pentamer is disrupted while it remains intact outside it (indicated in the figure). c, Interactions between subunits within the TrwI/VirB6 pentamer. Two adjacent subunits are shown in pink and red ribbon, respectively. Interface residues in the subunit in red are shown as surface coloured in grey. All secondary structures where residues are involved in interactions are labelled. The positions of the TMs as defined in the electron density map are shown. d, Interactions between subunits within the TrwJ/VirB5 pentamer. Two adjacent subunits are shown in orange and yellow ribbon, respectively. Interface residues in the subunit in orange are shown in surface representation coloured in orange. All secondary structures where residues are involved in interactions are labelled. e, Superposition of TrwJ/VirB5 with two known VirB5 homologues and one pore-forming protein Hemolysin E. Left: superposition with TraC (PDB entry code: 1R8I19), the VirB5 homologue of the pKM101 plasmid-encoded T4SS. Middle: superposition with CagL (PDB entry code: 3ZCI20), the VirB5 homologue in the Cag pathogenicity island of Helicobacter pylori. Right: superposition with Hemolysin E (PDB entry code: 6MRU68), a bacterial pore-forming protein, one of top hits in DALI69. The superposition is particularly good with the C-terminal half of CagL and Hemolysin E (RMSD of 3.1 and 3.2 Å in Cα atoms, respectively). f, Interactions between TrwJ/VirB5 and TrwI/VirB6. One TrwJ/VirB5 subunit (in orange ribbon) interacts with two TrwI/VirB6 subunits (shown in red and pink). For TrwI/VirB6, the regions of two subunits involved in interactions with TrwJ/VirB5 are shown as a surface while the rest of the molecules are shown in ribbon. Secondary structures contributing residues to the interfaces are labelled. g, Interactions between subunits within the TrwG/VirB8peri homo-trimeric unit. Left: the homo-trimeric TrwG/VirB8peri unit. Each subunit is shown in a different colour, pale cyan (MolC), pale green (MolB) and pale blue (MolA), respectively. Centre: the VirB8peri dimer from H. pylori21 (PDB entry code: 6IQT). The orientation shown results from a superposition of this dimer on MolA/MolB of TrwG/VirB8peri. As can be seen, the interface between subunits within this dimer is similar to that of the MolA/MolB interface between TrwG/VirB8peri subunits. Right: the VirB8peri dimer from Brucella suis22 (PDB entry code: 2BHM). The orientation shown results from a superposition of this dimer on MolB/MolC of TrwG/VirB8peri. As can be seen, the interface between subunits within this dimer is similar to that of the MolB/MolC interface between TrwG/VirB8peri subunits. RMSDs are reported in main text. Secondary structures contributing residues to the interfaces are labelled. h, Interface between TrwG/VirB8peri trimeric units in the Arches hexamer. In the T4SS structure presented here, six trimeric units come together to form the Arches. Inset: top view of the TrwG/VirB8peri trimeric units forming the Arches hexamer. The dashed lined box locates the region zoomed-in at right. One trimeric unit is colour-coded as in panel g, while the adjacent trimeric units are coloured in yellow orange. The secondary structures involved in interactions between trimeric units are labelled.