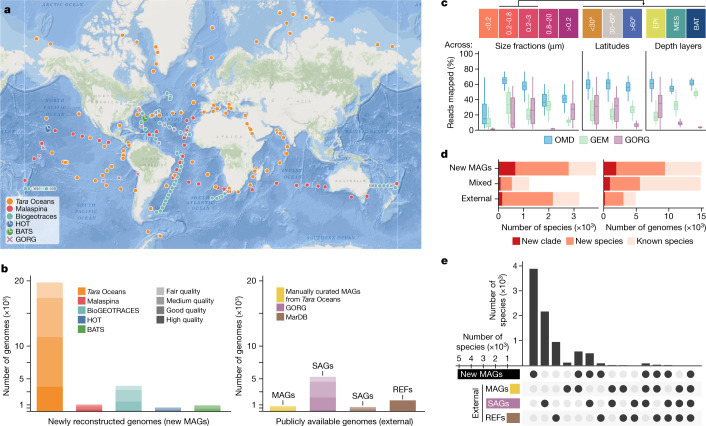
Fig. 1. Reconstruction of MAGs at the global scale fills gaps in ocean phylogenomic diversity.
a, A total of 1,038 publicly available ocean microbial community genomes (metagenomes) were collected at 215 globally distributed sites (between 62° S to 79° N and 179° W to 179° E). Map tiles © Esri. Sources: GEBCO, NOAA, CHS, OSU, UNH, CSUMB, National Geographic, DeLorme, NAVTEQ and Esri. b, These metagenomes were used to reconstruct MAGs (Methods and Supplementary Information), which varied in numbers and quality (Methods) across different datasets (colour coded). Reconstructed MAGs were complemented with publicly available (external) genomes, including manually curated MAGs26, SAGs27 and REFs. 27 to compile the OMD. c, The OMD improves the genomic representation (mapping rates of metagenomic reads; Methods) of ocean microbial communities by a factor of two to three compared with previous reports based solely on SAGs (GORG)20 or MAGs (GEM)16, with a more consistent representation across depth and latitudes. <0.2, n = 151; 0.2–0.8, n = 67; 0.2–3, n = 180; 0.8–20, n = 30; >0.2, n = 610; <30°, n = 132; 30–60°, n = 73; >60°, n = 42; EPI, n = 174; MES, n = 45; BAT, n = 28. d, Grouping the OMD into species-level (95% average nucleotide identity) clusters identified a total of around 8,300 species, over half of which were previously uncharacterized based on taxonomic annotations using the GTDB (release 89)13. e, A breakdown of the species by genome type reveals a high complementarity of MAGs, SAGs and REFs in capturing the phylogenomic diversity of the ocean microbiome. Specifically, 55%, 26% and 11% of the species were specific to MAGs, SAGs and REFs, respectively. BATS, Bermuda Atlantic Time-series; GEM, Genomes from Earth’s Microbiomes; GORG, Global Ocean Reference Genomes; HOT, Hawaiian Ocean Time-series.