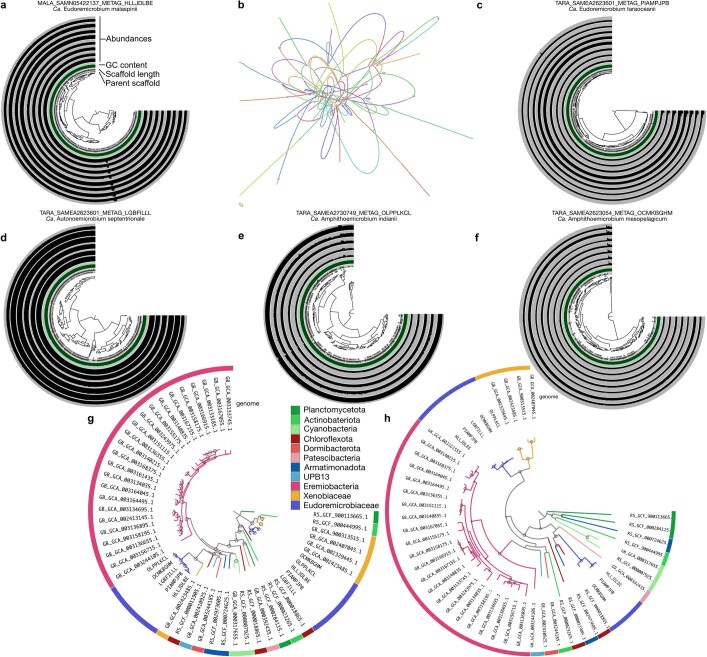
Extended Data Fig. 6. Manual inspection of Ca. Eudoremicrobiaceae MAGs and phylogeny of the duplicated marker gene COG0124.
(a, c–f) Anvi'o interface of representatives of the five Ca. Eudoremicrobiaceae species reveals stable abundance correlation patterns across the vast majority of the genomes, indicative of low contamination rates (Supplementary Information). (b) Inspection of the assembly graph for Ca. E. malaspinii (Supplementary Information) showed that all scaffolds from the representative genomes were connected with the exception of a single 20 kb one. (g) Investigating the evolutionary history of duplicated single-copy marker genes (here COG0124), we found consistent duplication across Ca. Eudoremicrobeaceae and the parent order UBP9, thus ruling out the duplication as a signal of contamination in the binning process. The different evolutionary history of the second copy of COG0124 (right-hand side of the tree), with closer relationship to Actinobacteria suggests that introgression events (including before the UBP9 and Ca. Eudoremicrobiaceae split) could be the origin of the increased genome size and biosynthetic potential observed in Ca. Eudoremicrobiaceae. (h) Similar patterns can be found in the second duplicated marker gene (COG0522), although duplication was not detected across all Ca. Eudoremicrobeaceae spp. representatives.