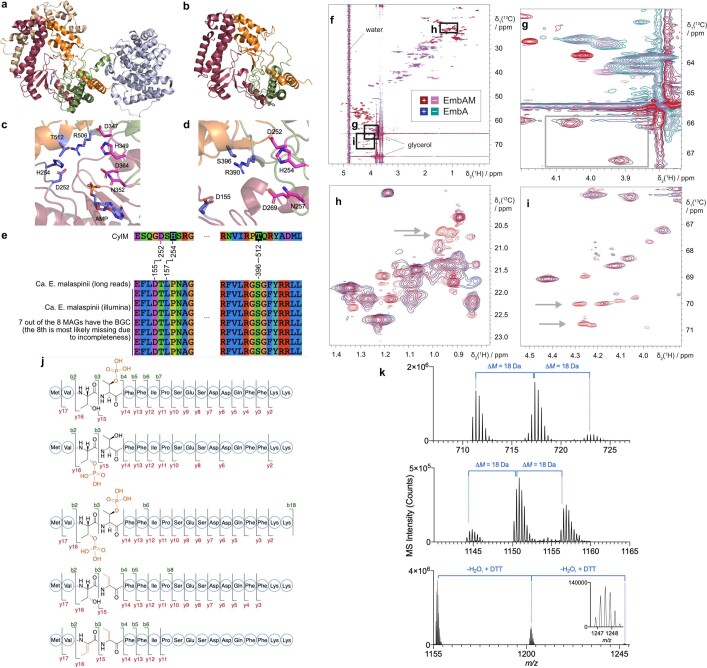
Extended Data Fig. 9. EmbM structural prediction and comparison to CylM (PDB: 5DZT); NMR and Mass spectrometry data for modified EmbA peptides.
(a) CylM crystal structure43. Coloured domains are involved in phosphorylation/dehydration and the domain in grey is responsible for cyclization. (b) EmbM structure prediction, highlighting similarities to CylM. (c) CylM active site. Residues in pink are proposed to be involved in phosphorylation and residues in purple are necessary for elimination. (d) Modelled active site of EmbM. (e) Multiple sequence alignment showing that mutated residues in the catalytic site are conserved across the independent Ca. E. malaspinii reconstructions. (f) Overlay of 2D [13C,1H] HSQC spectra of EmbA and modified EmbA (EmbAM). Multiplicity editing leads to positive signals for CH and CH3 groups (EmbA: blue, EmbAM: red) and negative signals for CH2 groups (EmbA: cyan, EmbAM: magenta). Regions of interest are identified with boxes and major buffer signals are labelled. (g) Serine Cβ region. Serine Cβ moieties are identified by the negative sign of the signal (CH2-group), and the average chemical shift of 63.8 ppm. A change of the Cβ chemical shift of typically +3 ppm is expected upon a phosphorylation event104, but there are no negative signals visible in the expected region in the EmbAM spectrum (grey box). (h) and (i): threonine Cγ and Cβ regions, respectively, as identified by chemical shift and sign of signals. In the EmbAM spectra, additional signals are visible at expected chemical shifts for phosphorylated threonine residues, i. e. at a 13C chemical shift of 20.5 ppm for Cγ (grey arrows in h) and 70 ppm for Cβ (grey arrows in i). (j) HR-MS/MS fragmentation of EmbA core at different modification stages (cleaved with LahT150). (k) Mass spectrum of dehydrated EmbA species: unmodified, single- and double dehydrated EmbA core (top); unmodified, single- and double dehydrated EmbA cleaved with trypsin (middle); and unmodified, single- and double dehydrated, DTT adduct of EmbA cleaved with trypsin (bottom).