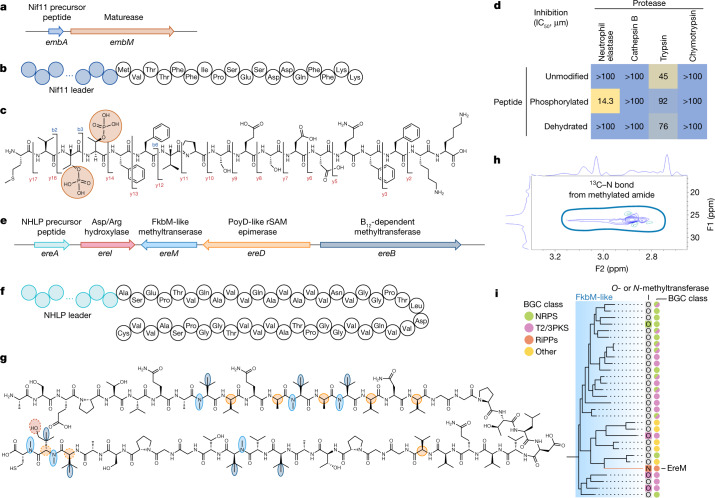
Fig. 4. ‘Ca. Eudoremicrobiaceae’ spp. are a source of unusual enzymology and natural product structure.
a–c, In vitro heterologous expression and in vitro enzyme assays of a novel (RefSeq = 0.29) RiPP biosynthetic cluster specific to the deep ocean species ‘Ca. E. malaspinii’ led to the production of a di-phosphorylated product. c, Modifications were identified using high-resolution (HR) MS/MS (fragmentation is indicated by the b and y ions on the chemical structure) and NMR (Extended Data Fig. 9). d, This phosphorylated peptide displayed low-micromolar mammalian neutrophil elastase inhibition, which was not found for the control and dehydrated peptides (dehydration induced by chemical elimination). The experiment was repeated three times, leading to similar outcomes. e–g, Heterologous expression of a second novel RefSeq = 0.33) proteusin biosynthetic cluster sheds light on the functionality of four maturases modifying a 46-amino-acid core peptide. Residues are coloured on the basis of predicted modification sites from HR-MS/MS, isotope labelling and NMR analyses (Supplementary Information). Dashed colouring indicates that the modification occurs on either of the two residues. The figure represents a compilation of numerous heterologous constructs to display the activity of all maturases on the same core. h, Inset of the NMR data of the backbone amide N-methylation. The complete results are shown in Extended Data Fig. 10. i, Phylogenetic placement of the FkbM maturase of the proteusin cluster among all FkbM domains found in the MIBiG 2.0 database revealed an enzyme of this family with N-methyltransferase activity (Supplementary Information). Schematic representations of the BGCs (a,e), the structure of the precursor peptides (b,f) and the proposed chemical structures of the natural products (c,g) are shown.