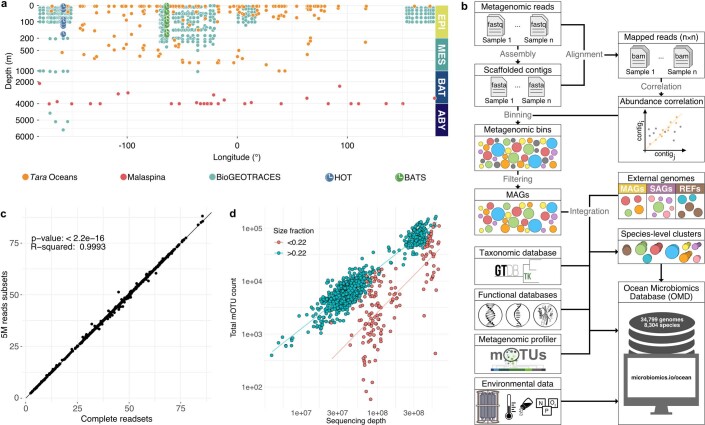
Extended Data Fig. 1. Depth distribution of the metagenomes used in this study; overview of the bioinformatic pipeline and proxies for sequencing depth.
(a) 1,038 publicly-available ocean microbial community genomes (metagenomes) were collected across all major depth layers (1 - 5,601 m) in the context of different ocean expeditions and time series programmes; EPI - epipelagic layer; MES - mesopelagic layer; BAT - bathypelagic layer; ABY - abyssopelagic layer. (b) Quality-controlled, high-throughput DNA sequencing reads from ocean microbial community samples were individually assembled into metagenomic scaffolded contigs (scaffolds). Sequencing reads from large subsets (n ranging from 58 to 610) of all samples were aligned to scaffolds of each individual sample to compute relative copy-number abundances for each scaffold in each sample. Based on a combination of tetranucleotide frequency, within-sample co-abundance and between-sample abundance correlations, scaffolds were grouped into a total of 62,874 metagenomic bins, each with total nucleotide sequence lengths of > 200 kb. These metagenomic bins were filtered for genome completeness and contamination, resulting in 26,293 metagenome assembled genomes (MAGs). These MAGs were complemented with external sets of MAGs, single amplified genomes (SAGs) and genomes from cultured isolates (REFs). The combined set of 34,799 genomes was clustered at the species level using a 95% average nucleotide identity (ANI) and, along with taxonomic and functional annotations, abundance profiles and contextual information, compiled into the Ocean Microbiomics Database (OMD); see methods for details (Methods). (c) Comparing mapping rates obtained from mapping subsampled readsets compared to those obtained from mapping the total number of reads shows that this procedure yields almost identical results at considerably less computational costs. (d) mOTUs counts as a good proxy for sequencing depth. We find a strong correlation in prokaryote-, particle-enriched and virus-depleted communities, while this correlation is more variable in virus-enriched communities. This observation is actually in support of using the mOTUs count rather than sequencing depth when focusing on the bacteria and archaeal component of microbial communities, as we do here.