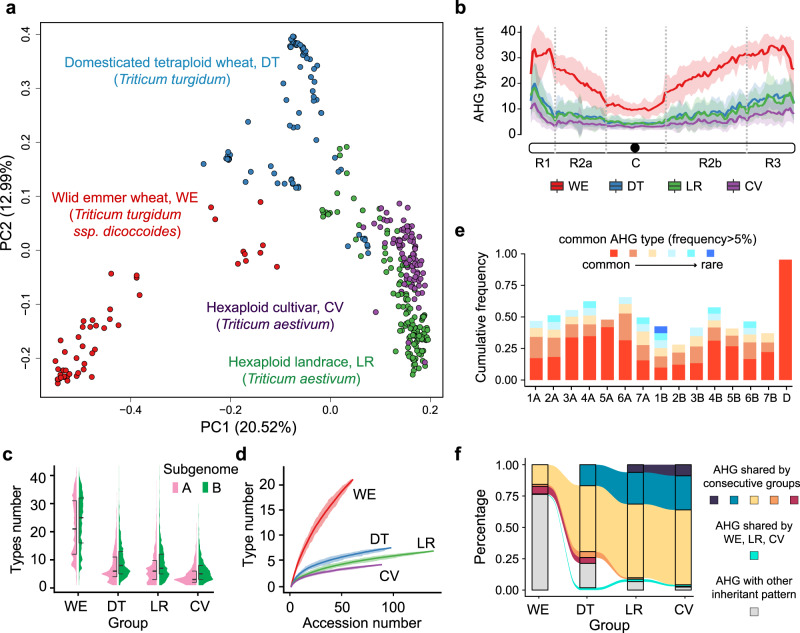
Fig. 2. AHG reveals the transition of gene pools in polyploid wheat.
a Principal component analysis plot constructed with AHG-based distances for all 5-Mb windows in A&B genomes. b Counts of AHG types across chromosome model of A&B subgenomes. Solid line and colored area, mean±sd. WE, wild emmer wheat. DT, domesticated tetraploid wheat. LR, hexaploid wheat landrace. CV, hexaploid wheat cultivar. The chromosomal zones R1, R2a, C, R2b and R3 are consistent with previous publication32. c Violin plot showing the count distributions of AHG types in the A&B subgenomes for each group. d Saturation curves showing the cumulative number of AHG types versus the number of accessions included. Solid line and shaded region, average and 90% confidence intervals for 14 A&B chromosomes. e Stacked bar graph showing the frequency and corresponding order of common AHGs (frequency > 5%) by chromosome. “D” indicates the average value of 7 chromosomes in D subgenome. f Proportion of shared AHGs among four taxonomic groups shown by alluvial plot revealed genetic flows through the route of WE-DT-LR-CV. A dramatically higher proportion of AHGs in the hexaploid groups can be traced back to the DT group than to the WE group using the collected samples, indicating DT as the closest gene pool for hexaploid wheat. Source data are provided as a Source Data file.