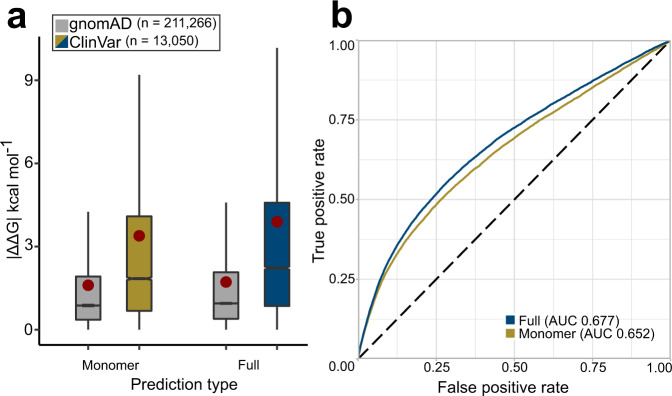
Fig. 1. Using full protein complex structures improves discrimination between pathogenic and putatively benign missense variants.
a Comparison of predicted | ΔΔG | values for putatively benign gnomAD and pathogenic ClinVar mutations when calculated using monomer structures only, or by also employing full biological assemblies and therefore also considering intermolecular interactions. Boxes denote data within 25th and 75th percentiles, and contain median (middle line) and mean (red dot) value notations. Whiskers extend from the box to furthest values within 1.5x the inter-quartile range. All pairwise group comparisons (both between and within prediction types) are significant (p < 2.5 × 10−28, two-sided Dunn’s test for multiple pairwise comparisons, corrected for multiple testing by Holm’s method; see ‘Methods’). b Receiver operating characteristic (ROC) curves and area under the curve (AUC) for discrimination between ClinVar and gnomAD mutations using monomer and full | ΔΔG | values. Source data are provided as a Source Data file.