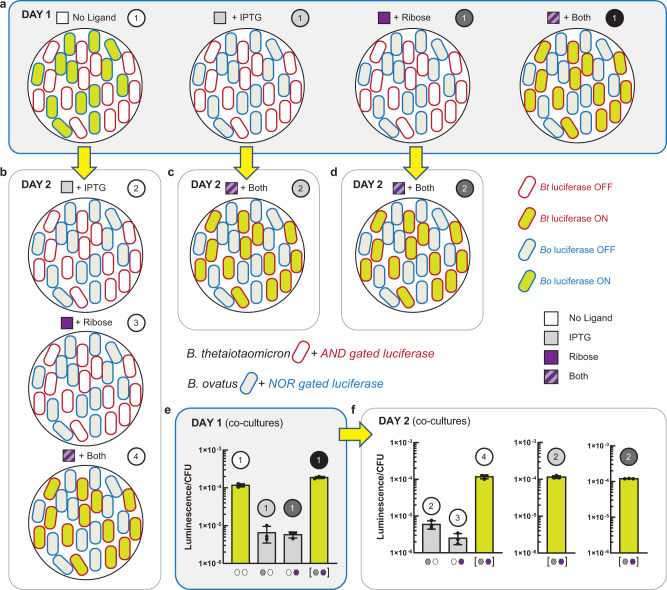
Fig. 7. Sequential programming in co-culture.
a Co-culture composed of B. thetaiotaomicron and B. ovatus harboring different logic gates. B. thetaiotaomicron integrated with an AND gate was grown in the same TYG culture as B. ovatus integrated with a NOR gate (both gates regulating luciferase expression). Co-cultures were grown in the presence of all combinations of inducers (each at a final concentration of 10 mM) yielding four total cultures which were independently assayed for luciferase activity (Methods). b Sequential programs derived from the “No Ligand” culture in a. The initial co-culture was used to seed fresh media containing additional inducers (i.e., IPTG, ribose, or both ligands). These new co-cultures were grown and assayed for luciferase activity (f, left bar chart). c Sequential program derived from the “+ IPTG” culture in a. The initial co-culture was used to seed fresh medium containing both ligands. This new co-culture was subsequently assayed for luciferase activity (f, middle bar chart). d Sequential program derived from the “+ Ribose” culture in a. The initial co-culture was used to seed fresh medium containing both ligands. This new co-culture was subsequently assayed for luciferase activity (f, right bar chart). e Luciferase activity of co-cultures on day 1. Numbered circles correspond to the co-cultures illustrated in a. f Luciferase activity of co-cultures on day 2. Numbered circles correspond to the co-cultures illustrated in b–d. Source data are provided as a Source Data file. Data represent the average of n = 3 biological replicates. Error bars correspond to the SEM of these measurements.