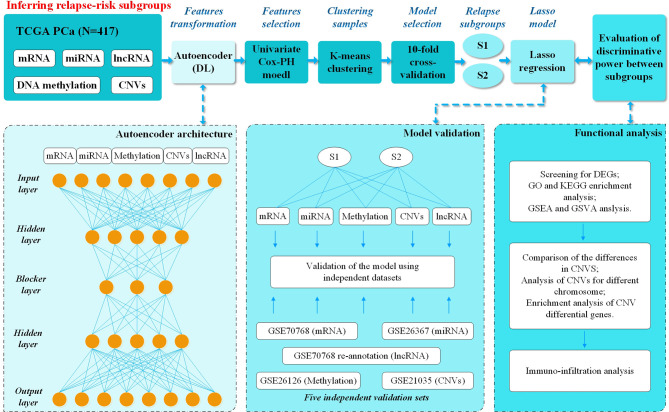
Figure 1.
Overall workflow. Firstly, mRNA, miRNA, DNA methylation, CNVs, and lncRNA deep features from the TCGA PCa cohort were stacked up as input features for the autoencoder, a deep-learning method. Then each of the new and transformed features in the bottleneck layer of the autoencoder were subjected to univariate Cox-PH models to select the features associated with relapse. Then K-mean clustering was applied to relapse-associated deep features and 10-fold cross-validation was applied to analyze the C-index for different clusters and relapse in 8 DL models. The best model (model_3) with better discriminative ability was finally selected using Kaplan-Meier plotter between two models with the highest C-index. Then the Lasso method was used to filter out the relapse-associated feature labels, according to model_3 subgroups, from the database of TCGA, including mRNA, miRNA, DNA methylation, CNVs and lncRNA. The Lasso model was constructed, and five external validation sets from GEO were used to evaluate its prediction ability. Last but not least, functional analysis was performed to understand the different characteristics between two relapse-associated subgroups.