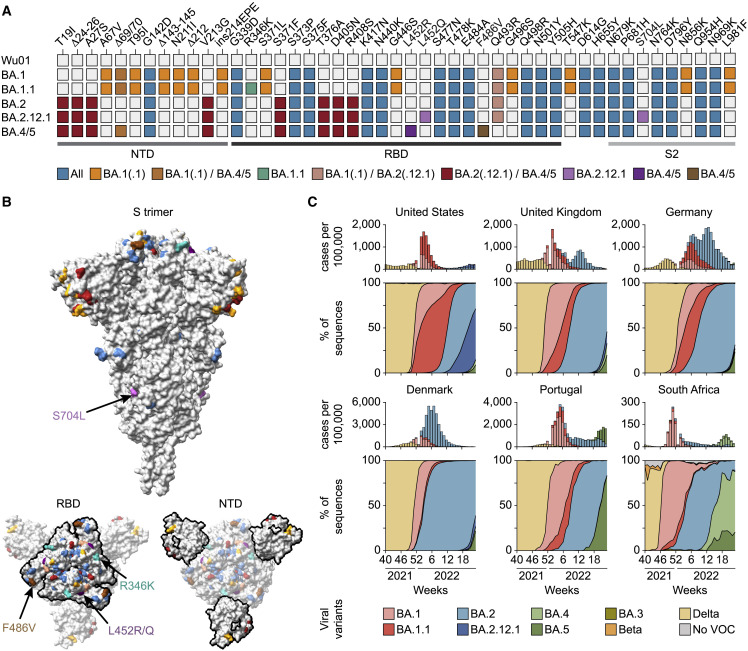
Figure 1.
Omicron sublineage differences
(A) Spike amino acid changes in Omicron sublineages relative to Wu01.
(B) Locations of Omicron-sublineage amino acid changes on the SARS-CoV-2 spike (PDB: 6XR8) with colors as in (A), and RBD and NTD outlined in the bottom models. Residues with mutations exclusive to individual sublineages are indicated by arrows.
(C) Top panels indicate weekly reported SARS-CoV-2 infections as aggregated by the Johns Hopkins University CSSE COVID-19 data repository and Our World in Data, with variant proportions extrapolated from weekly GISAID SARS-CoV-2 database variant sequences (accessed on June 20, 2022) shown in bottom panels.
NTD, N-terminal domain; RBD, receptor-binding domain; PDB, Protein Data Bank.