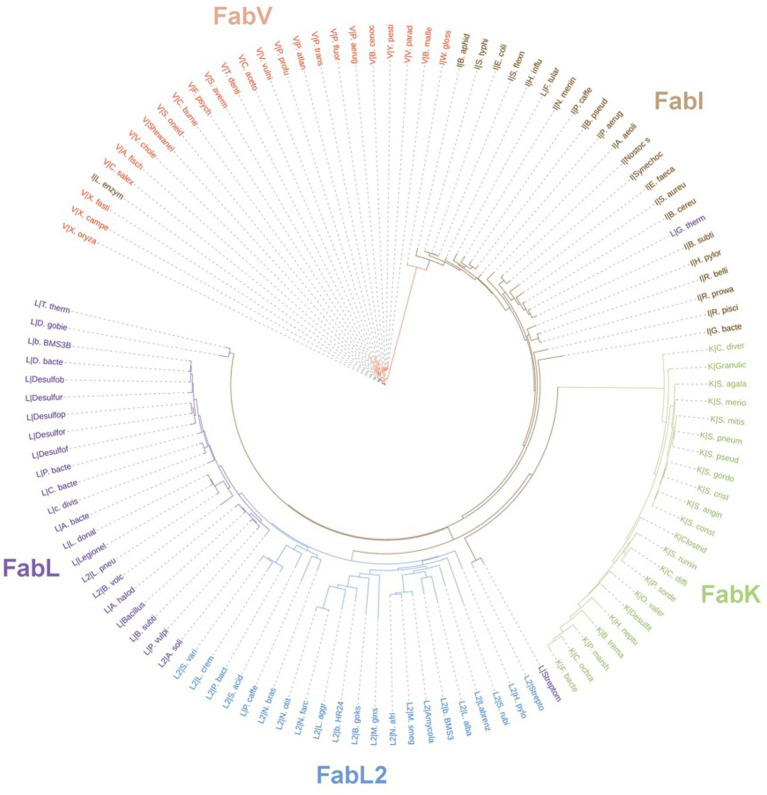
Figure 13.
Unrooted phylogenetic tree inferred from the sequences of ENRs from the classes FabK, FabL, FabL2, FabI, and FabV. Branch lengths denote amino acid substitutions for that node. The letters behind a “|” symbol refer to their respective ENR class. The Uniprot identifiers for each one of the species present in the tree are listed on Supplementary Table 1. Sequences were downloaded from Uniprot and aligned using the multiple alignment tool ClustalW (Larkin et al., 2007). The resulting phylip alignment file was used as input for RAxML (Stamatakis, 2014) and the best substitution model was chosen automatically. Trees were visualized and customized in the webserver of iTOL (Letunic and Bork, 2021).