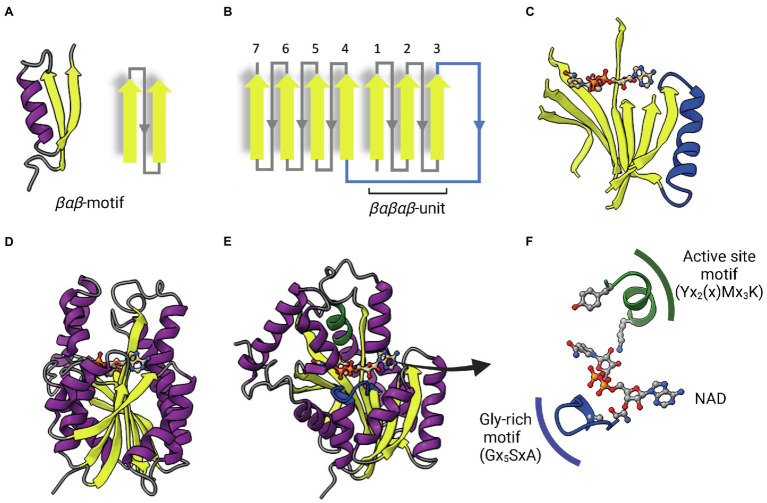
Figure 2.
The dinucleotide-binding Rossmann fold. (A) A representative βαβ-motif and its topological representation. An α-helix (purple) connects two parallel β-strands (yellow). (B) Topological representation of the Rossmann fold. The βαβ-motifs form two sets of βαβαβ units that are connected by a long α-helix that function as a crossroad (in blue). (C) Representative parallel β-sheet of a Rossmann fold. The α-helix connecting the two sets of βαβαβ units through strands 3 and 4 is represented in blue. The NAD cofactor is also represented to indicate that the cofactor-binding site lies above this central parallel β-sheet. (D) Structure of InhA (PDB: 1ENY). The central parallel β-sheet is flanked by two sets of α-helices on each side. (E) InhA (PDB: 1ENY) with the central parallel β-sheet in lateral view. The active site motif Yx2(x)Mx3K and the cofactor-binding glycine-rich motif (Gx5SxA) are shown in green and blue, respectively. (F) Detailed view of NAD, the active site and Glycine-rich motifs. The NAD cofactor, the first glycine from the Gx5SxA motif (G14), catalytic tyrosine (Y158) and lysine (K165) from the signature motif Yx2(x)Mx3K are represented as Ball & Stick atomic models. Created with UCSF ChimeraX (Pettersen et al., 2021) and BioRender.com.