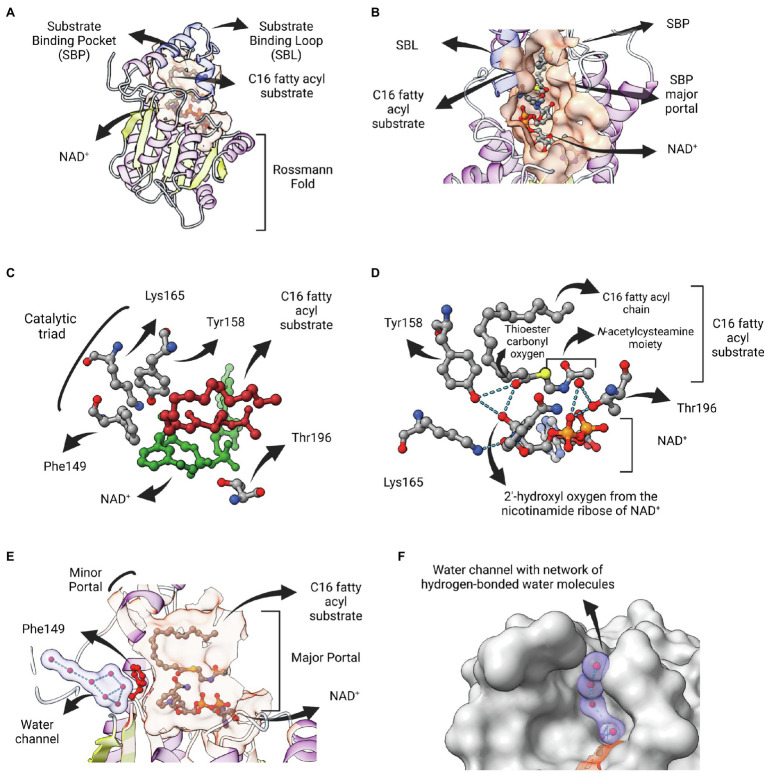
Figure 5.
InhA bound to NAD and fatty acyl substrate. (A) Structure of the ternary complex of Mycobacterium tuberculosis InhA bound to NAD+ and a C16 fatty acyl substrate (PDB: 1BVR). Both cofactor-binding site and fatty acyl binding site lie within the same structural substrate binding pocket (SBP; beige surface). The substrate-binding loop (SBL) of mycobacterial FabIs like M. tuberculosis InhA (residues 196–219, in blue) are longer than their bacterial orthologues, presumably to accommodate longer fatty acyl substrates. The fatty acyl substrate can be viewed in its U-shaped conformation. The Rossmann fold is also represented. (B) View of the substrate binding pocket from the major portal. The U-shaped fatty acyl chain lies just ahead of the cofactor. (C) Detailed view of the catalytic triad (F149, Y158 and K165), together with T196, fatty acyl substrate and cofactor. (D) Hydrogen bond interactions among the catalytic Y158 and K165, the fatty acyl substrate and the NAD cofactor. (E) Lateral view of the substrate binding pocket of InhA displaying a network of water molecules inside a water channel. The residue F149 presumably gates the access of this water channel to the active site of InhA. This network of water molecules was proposed to be implicated in the protonation of the enolate intermediate. The minor portal is also indicated. (F) Surface view of InhA with the external part of the hydrogen-bonded network of water molecules inside the water channel. Binding pocket was identified using CASTp 3.0 (Tian et al., 2018). Created with UCSF ChimeraX (Pettersen et al., 2021) and BioRender.com.