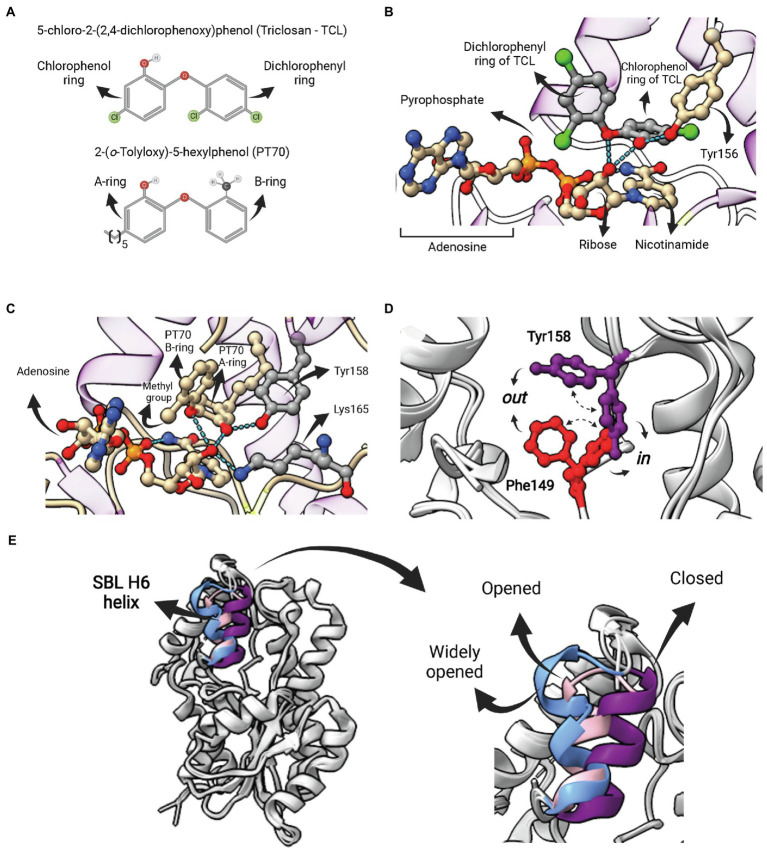
Figure 7.
Competitive inhibitors of FabI—Triclosan and PT70. (A) Structure of 5-chloro-2-(2,4-dichlorophenoxy)phenol (Triclosan—TCL) and the TCL derivative 2-(o-Tolyloxy)-5-hexyphenol (PT70). (B) Ternary complex of EcFabI bound to TCL and NAD (PDB:1QSG). Hydrogen bonds between the ether oxygen and the chlorophenol ring hydroxyl oxygen with the 2′-oxygen from the nicotinamide ribose of NAD are highlighted. (C) Ternary complex of InhA bound to NAD+ and PT70 (PDB: 2×23). Hydrogen bonds between the inhibitory compound and the catalytic Y158 and K165 residues are highlighted. This compound also makes hydrogen bonds with the 2′-nicotinamide ribose oxygen of NAD+. (D) Superposition of two InhA structures highlighting to possible conformations for F149 and Y158. In both cases, the in conformation is the one in which the residue points toward the active site, as opposed to the out conformation. In one structure (PDB: 4D0S) the F149 is in the in conformation, while Y158 is in out conformation. The second structure (PDB: 3FNH) the situation is the opposite (F149 out, Y158 in). Note that the configuration (F149 in, Y158 in) is not allowed due to steric hindrance. (E) Structural flexibility of the substrate-binding loop (SBL) helix 6 (residues 196–206) from InhA. This region is found in variable conformations in different structures, but they can be grouped in four types, each of them represented by one structure in the picture: disordered (4TRM), widely opened (1P44), open (2AQ8) and closed (3FNH). Potent slow-onset inhibitors usually induce the ordering of the SBL H6 helix into the closed conformation. The superposition of structures was performed using the Matchmaker tool from UCSF ChimeraX. Created with UCSF ChimeraX (Pettersen et al., 2021) and BioRender.com.