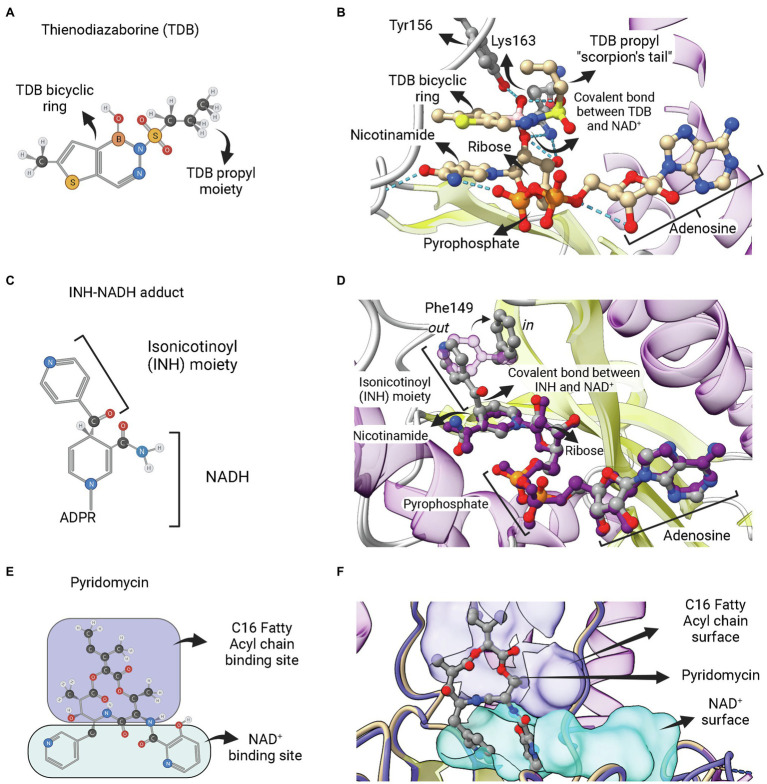
Figure 8.
Adduct forming compounds and a bisubstrate inhibitor. (A) Structure of a diazaborine derivative (thienodiazaborine—TDB). (B) Ternary complex of EcFabI bound to NAD+ and TDB (PDB: 1DFH). The TDB propyl moiety turns back in a conformation reminiscent to a scorpion’s tail (Baldock et al., 1996). The covalent bond between TDB and NAD+ is indicated. (C) Structure of the INH-NADH adduct. Only the nicotinamide portion of NADH is represented. (D) Comparison of the complex of InhA bound to the INH-NAD adduct (PDB: 1ZID) with the binary complex of InhA with NAD+ (PDB: 2AQ8). Both structures almost completely overlap, except for the conformation of F149, which rotates from the out conformation in the binary complex InhA-NAD+ to the in conformation in the enzyme complex with INH-NADH. This rotation is required to accommodate the isonicotinoyl (INH) moiety of the INH-NADH adduct. (E) Structure of the pyridomycin bisubstrate inhibitor. The portions that occupies the fatty acyl chain binding site and the NAD+ binding site are indicated. (F) Binary complex of InhA bound to pyridomycin (PDB: 4BII). The superposition of structures was performed using the Matchmaker tool from UCSF ChimeraX. Created with UCSF ChimeraX (Pettersen et al., 2021) and BioRender.com.