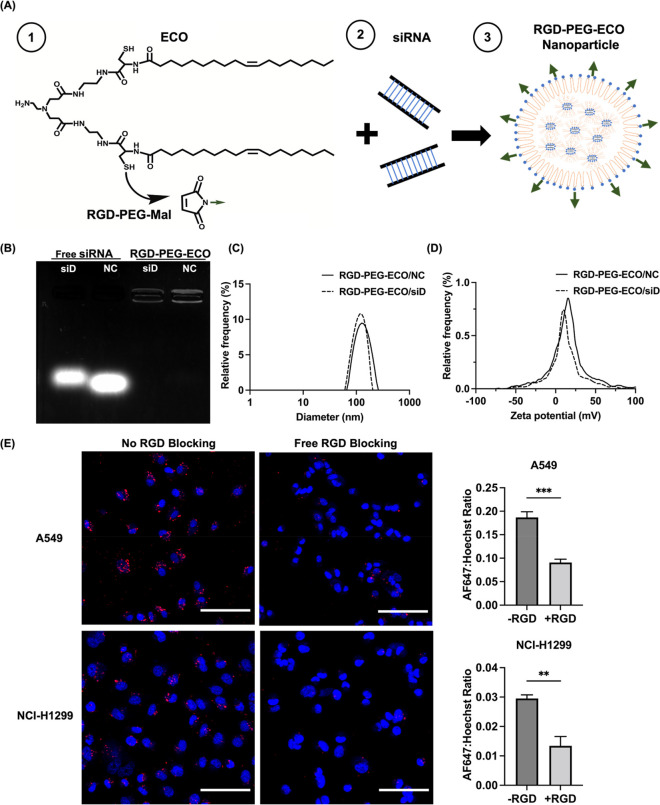
Figure 1.
Formation and characterization of RGD-PEG-ECO/siRNA nanoparticles. (A) Structure of ECO and scheme of RGD-PEG-ECO/siRNA formation. (B) Gel encapsulation assay shows strong entrapment of siDANCR and siNS in RGD-PEG-ECO nanoparticles with free siRNA as controls. RGD-PEG-ECO nanoparticles have uniform size (C) and zeta potential distributions (D) as measured with dynamic light scattering. The measured size is 119 nm for RGD-PEG-ECO/siDANCR nanoparticles and 116 nm for RGD-PEG-ECO/siNS nanoparticles; zeta potential is ca. +14 and +13 mV, respectively. (E) Confocal images (20× magnification) taken after 24 h blocking with excess free RGD peptide show reduced RGD-PEG-ECO nanoparticle uptake. Ratio of AF647 to Hoechst 33342 signal intensity is higher when no free RGD peptide is present in the transfection media (−RGD) compared to when excess RGD peptide is added (+RGD). Red: AF647; blue: Hoechst 33342 (n = 3). NC represents the negative control siRNA, siNS. siD represents siDANCR. Error bars denote standard error of measure; scale bars in all panels = 100 μm. ** p < 0.01, and *** p < 0.005 using unpaired t-test.