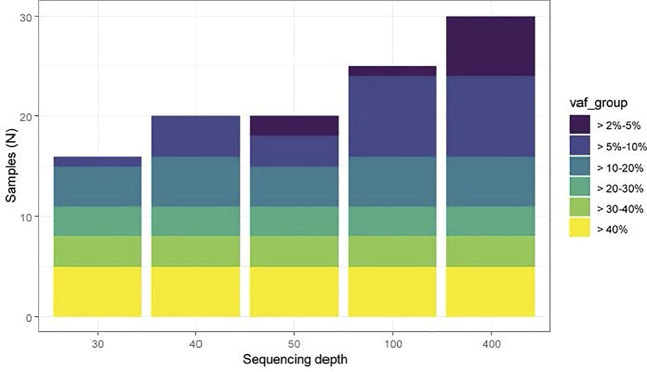
FIGURE 3.
Sensitivity of CHIP detection at different sequencing depths (Bick et al., 2020a). A set of 30 samples from a previously published CHIP cohort were computationally down sampled to 30x, 40x, 50x, 100x and 400x sequencing depth. TOPMed WGS data was typically in the 40x depth range across CHIP genes. WGS data has excellent sensitivity to detect CHIP clones with VAF >10%, and ∼50% sensitivity to detect CHIP VAF 5–10%, with minimal ability to detect CHIP clones <5%.