Fig. 1.
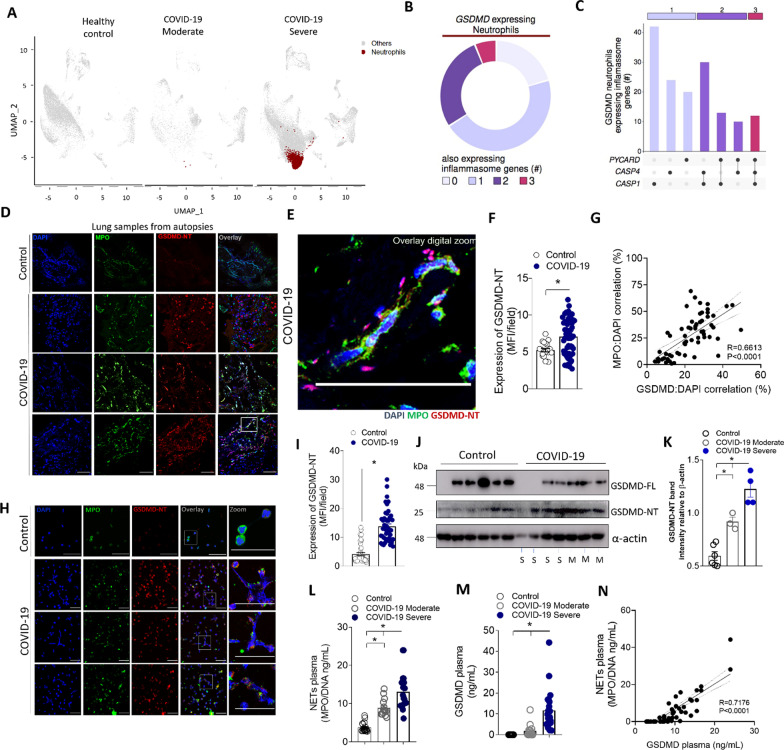
Neutrophils from COVID-19 patients undergoing NETosis express GSDMD. A Single-cell analysis of BALF from patients with COVID-19 across severity status (Healthy control, Moderate, and Severe). UMAP visualization from gene expression data of 66,452 cells, highlighting neutrophil expression cluster in severe COVID-19 patients (red) from bronchoalveolar lavage fluid (BALF) cells. B Pie chart plot representing the proportion of GSDMD expressing neutrophils. C UpSet plot showing the intersection of inflammasome genes expressed in neutrophils, including PYCARD, CASP4, and CASP1, derived from COVID-19 severe patients. The point diagram indicates the intersection among the genes and the bar graph shows the number of GSDMD expressing neutrophils in each intersection (y-axis). D Representative confocal analysis of GSDMD-NT and NETs in the lung tissue sample from autopsies of COVID-19 patients (n = 6 or control n = 3). Immunostaining for DNA (DAPI, blue), myeloperoxidase (MPO, green), and the GSDMD cleaved fraction (GSDMD-NT, red) are shown. The scale bar indicates 50 μm at 630× magnification. E Zoomed images of Fig. 1D inset white square. F The expression of GSDMD-NT was quantified by MFI per field. G Correlation between colocalization GSDMD-NT:DAPI and NETs (MPO:DAPI). H Representative confocal analysis of GSDMD and NETs in the blood neutrophils isolated from COVID-19 patients (n = 5) or controls (n = 5). Cells were stained for DNA (DAPI, blue), MPO (green), and GSDMD-NT (red). Scale bar indicates 50 μm, 4× digital zoom was performed in the inset white square. I Expression of GSDMD-NT was quantified by MFI per field. J Expression of full-length GSDMD (GSDMD-FL) and active GSDMD (GSDMD-NT) by Western blot. Moderate COVID-19 (M, n = 3) severe COVID-19 (S, n = 4), and healthy controls (n = 36). (K) Western blot quantification by densitometry. GSDMD-NT values obtained were normalized to total beta-actin (L) Circulating amounts of MPO/DNA-NETs and M GSDMD from plasma of patients with moderate COVID-19 (n = 15) severe COVID-19 (n = 21), and healthy controls (n = 320). N Correlation between plasmatic levels of MPO/DNA-NETs and GSDMD. The data are expressed as mean ± SEM (*p < 0.05; t test in F and I, Pearson’s correlation in G and M, one-way ANOVA followed by Tukey’s in K and L)