Figure 1. Splicing factors negatively autoregulate their own synthesis by promoting unproductive splicing of their own transcripts and also operate under variable loads of substrate pre-mRNA produced by other genes.
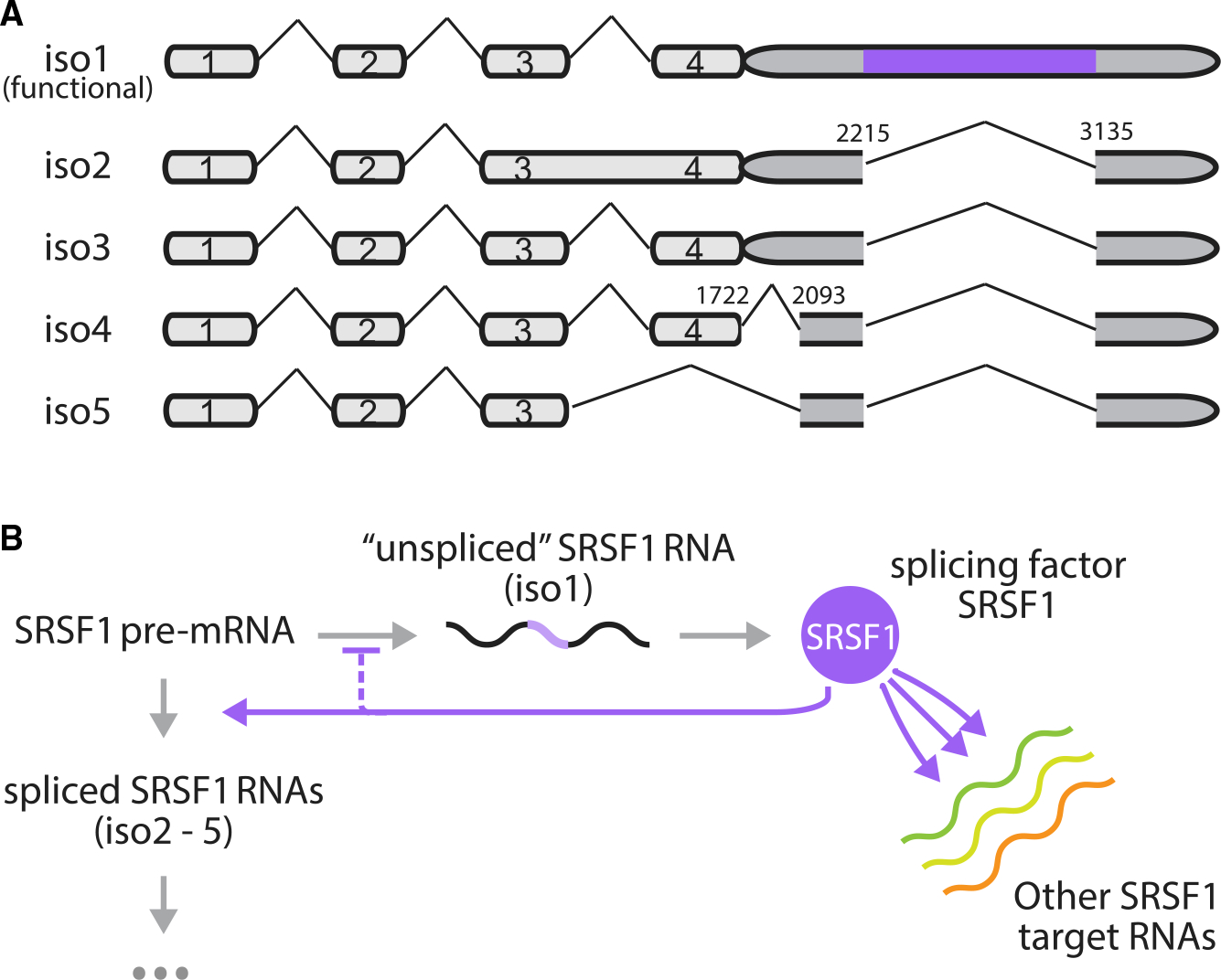
(A) Five isoforms of SRSF1 were observed in HEK293 cells. Each pre-mRNA molecule can be spliced to remove introns in the 3′ UTR (without light purple isoforms, isoforms 2–−5) or left unspliced (with light purple isoforms, isoform 1). Intron removal can lead to degradation through RNA surveillance pathways, while transcripts with retained introns in the 3′ UTR (isoform 1) are translated to produce more splicing factor SRSF1.
(B) Feedback occurs when splicing factors enhance intron removal from their own pre-mRNA, thus negatively regulating their own expression. Apart from their own transcripts, splicing factors additionally act on transcripts produced by other genes. The relative abundance of substrate pre-mRNAs can affect the allocation of splicing factors among transcripts, thereby influencing the dynamics of splicing negative autoregulation.
