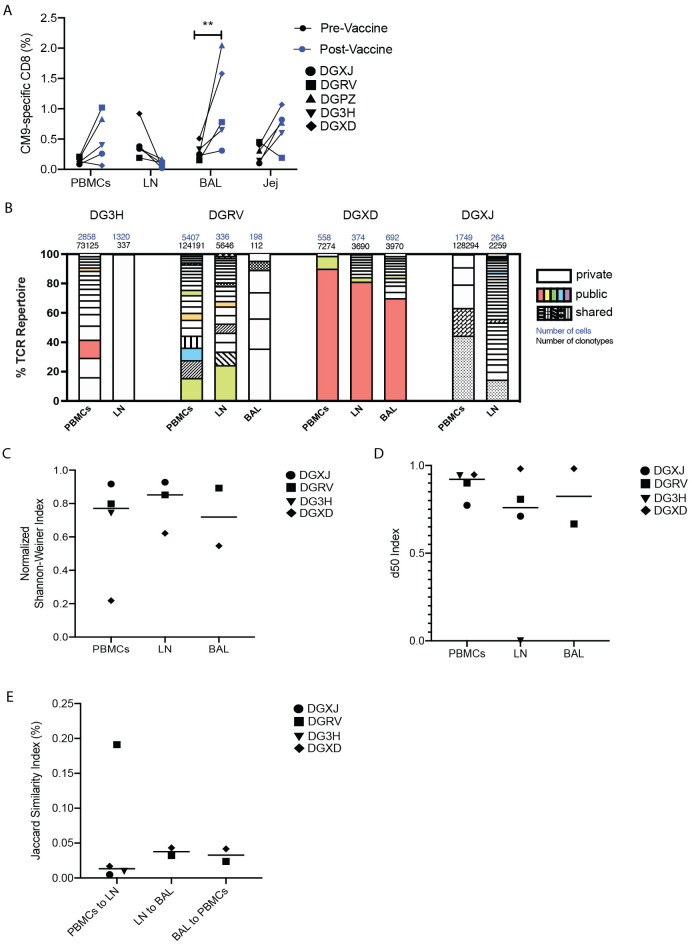
Fig 3. SIV-gag DNA vaccine induced a diverse TCR repertoire and tissue-specific clonotypes.
PBMCs, BAL, LN, and jejunum (jej) biopsies were sampled from rhesus macaques who had been administered with SIV-gag DNA vaccine. (A) CM9-specific CD8+ T cells in all tissues pre- and post- vaccine administration as a percentage of total CD8+ cells. (B) Clonotypes consisting of more than 1% of the TCR repertoire are represented as percentage of the TCR repertoire. “Public” clonotypes are the same clonotype (same V and J segments and same CDR3 amino acid sequence) found in multiple animals in this study, and matching clonotypes previously identified in the VDJdb database. “Shared” clonotypes were those found only in one animal but observed in multiple tissues. “Private” clonotypes were identified in a single anatomical site in a single animal. Total cell and TCR sequences numbers are listed above each column. (C) The normalized Shannon-Weiner diversity index for the TCR repertoires of all tissues. (D) The d50 diversity index for the TCR repertoires of all tissues. (E) The Jaccard similarity index for comparisons between each tissue’s TCR repertoire. In (C) to (E), data are presented as mean, with individual data points shown. Two-way ANOVA (A) and one-way ANOVA (C-E) were used to determine statistical significance. n = 5 animals.