Figure 4.
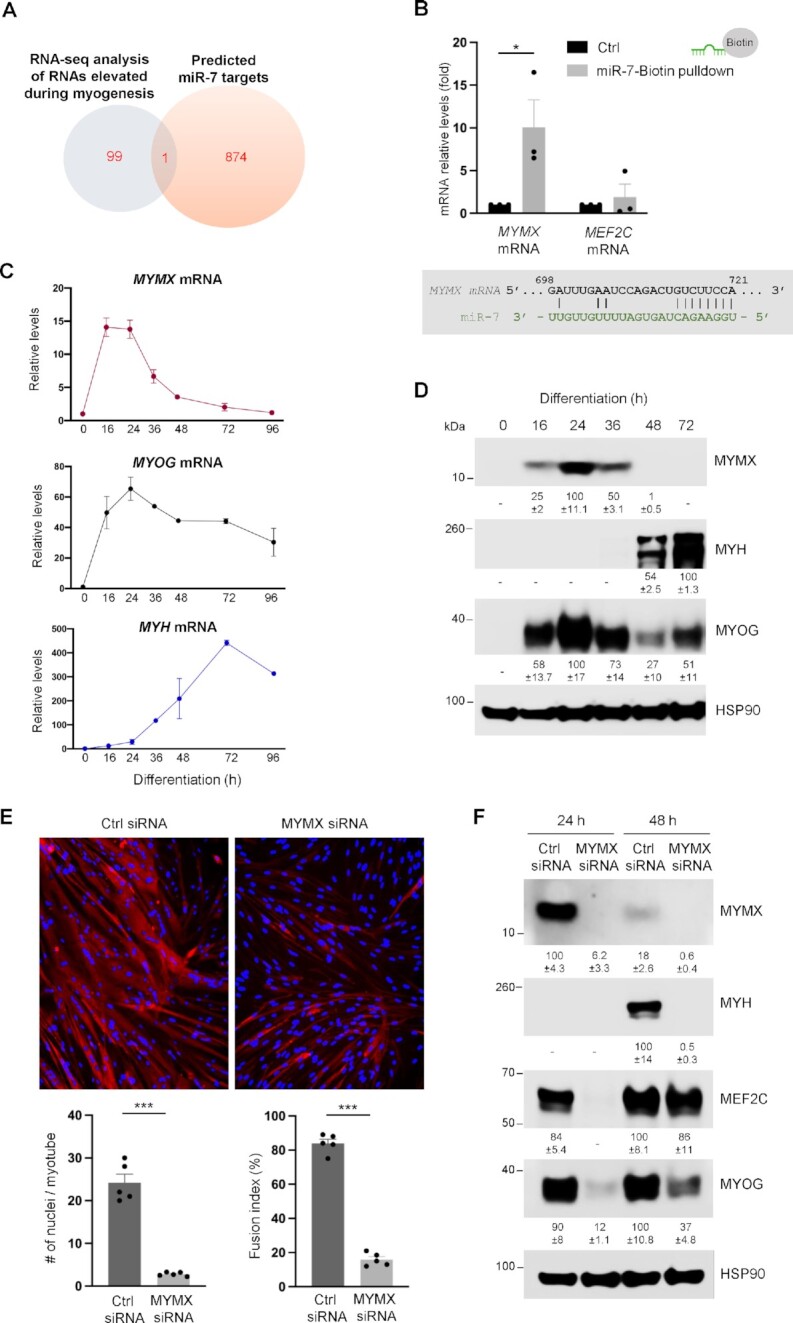
MYMX is a novel target of miR-7. (A) Venn diagram of RBPs from the top 100 upregulated RNAs in myogenesis by RNA-seq analysis (see the ‘Materials and Methods’ section) (gray), and the 875 predicted mRNA targets of miR-7 (orange); one mRNA was found in the intersection, MYMX mRNA. (B) AB678 myoblasts were transfected with biotinylated Ctrl miR or biotinylated miR-7; 6 h later, they were placed in differentiation medium for 12 h, harvested and RNA complexes were pulled down using streptavidin beads (see the ‘Materials and Methods’ section). The presence of potential miR-7 target mRNAs in the pulldown material was assessed by RT-qPCR analysis. The levels of RNAs tested in the pulldown were normalized to the levels of GAPDH mRNA. At the times indicated in differentiating AB678 cultures, the relative levels of MYMX mRNA and myogenic mRNAs were detected by RT-qPCR analysis (C), and the levels of MYMX and myogenic proteins were assessed by western blot analysis (D). (E) AB678 myoblasts were transfected with Ctrl siRNA or MYMX-directed siRNA; 24 h later, they were placed in differentiation medium for 72 h, and differentiation was monitored by assessing MYH levels by immunofluorescence (top), and the numbers of nuclei per myotube and fusion indices were quantified after assessing five separate fields per experiment (bottom). (F) As described in panel (E), the levels of MYMX and myogenic proteins were assessed by western blot analysis. Data in panels (B) and (D)–(F) represent the means ± SEM from three or more independent experiments. Significance was established using Student’s t-test. *P < 0.05; **P < 0.01; ***P < 0.001. Other data are representative of three or more biological replicates. In panels (D) and (F), bands were quantified by densitometry and the relative intensities are indicated.
