Figure 6.
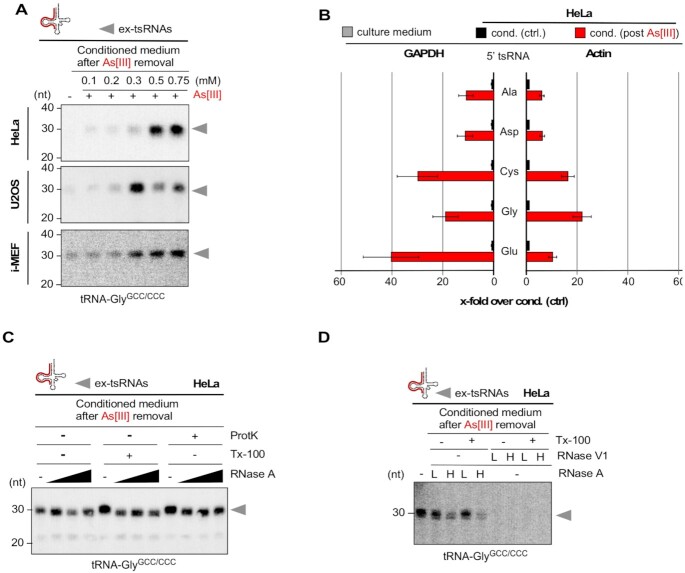
(A) Northern blotting of RNAs extracted from cell culture medium that was conditioned by HeLa, U2OS cells (0.2 ml medium) or i-MEF (0.4 ml medium), which had been exposed (for 1 h) to high micromolar concentrations of As[III], followed by culturing cells for 24 h after the removal of As[III] using a probe against the 5′ end of tRNA-GlyGCC/CCC. grey arrowheads: ex-tsRNAs. (B) qRT-PCR quantification of 5′ tsRNAs contained in RNAs extracted from cell culture medium (0.4 ml) that was either conditioned by HeLa cells, which had been exposed (for 1 h) to As[III] (0.75 mM), followed by culturing cells for 24 h after the removal of As[III] or by HeLa cells that were not exposed to As[III]. Bar-chart depicts the fold-change of specific 5′ tsRNAs in media (cond. post As[III]) over 5′ tsRNAs in control media (cond. ctrl), which were normalized to GAPDH (left) or ACTIN (right). Error bars depict standard error of the mean (SEM) of triplicate measurements from one experiment. (C) Northern blotting of RNAs extracted from HeLa cell culture medium (0.2 ml) of time-limited exposure (1 h) to As[III] (0.75 mM), followed by culturing cells for 24 h after the removal of As[III], and after being treated (either alone or in combination) with proteinase K (ProtK), detergent (Tx-100) and increasing concentrations of RNase A, using a probe against the 5′ end of tRNA-GlyGCC/CCC. grey arrowheads: ex-tsRNAs. (D) Northern blotting of RNAs extracted from HeLa cells as in (C), and after being treated (either alone or in combination) with detergent (Tx-100) and two concentrations (L, low; H, high) of either RNase A or RNase V1, using a probe against the 5′ end of tRNA-GlyGCC/CCC. Grey arrowheads: ex-tsRNAs.