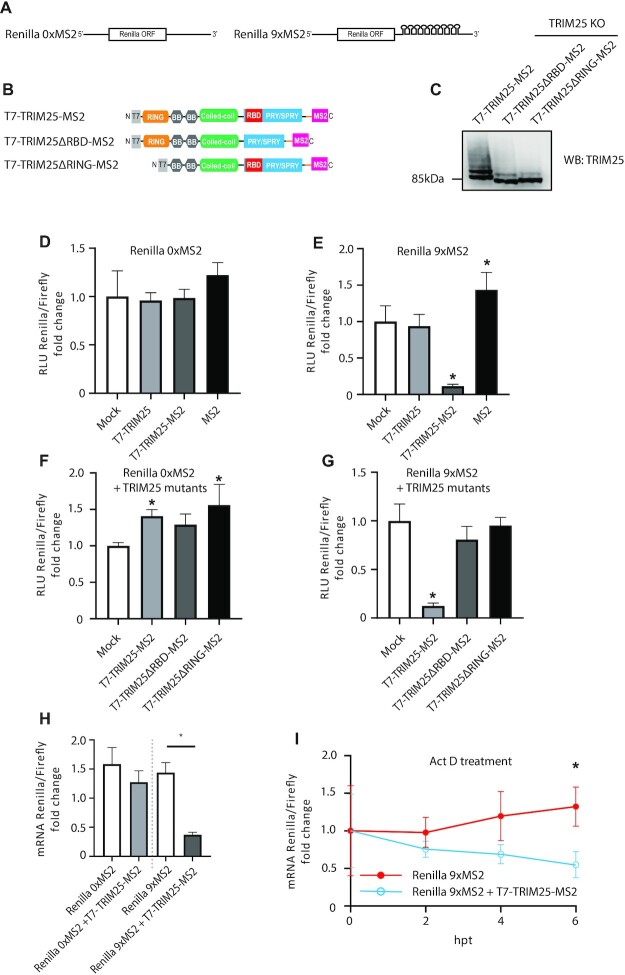
Figure 8.
Tethering of TRIM25-MS2 to MS2 stem loops triggers mRNA degradation. (A) Sequence architecture of Renilla mRNA with and without nine MS2 stem loops located in the 3′UTR. (B) Domain architecture of fusion protein of TRIM25, TRIM25ΔRBD or TRIM25ΔRING with MS2 domain (which binds MS2 RNA stem loops), tagged with T7 epitope. (C) T7-TRIM25-MS2, T7-TRIM25ΔRBD-MS2 or T7-TRIM25ΔRING-MS2 were overexpressed in HEK293 TRIM25 KO cells and levels of the proteins were compared to WT cells by western blot. (D-E) The psiCHECK2 plasmids without or with nine MS2 stem loops were co-transfected with plasmids expressing T7-TRIM25 WT, T7-TRIM25-MS2 fusion protein or MS2 on its own in to HEK293 TRIM25 KO cells and analysed with dual luciferase assay. (F, G) T7-TRIM25-MS2, T7-TRIM25ΔRBD-MS2 and T7-TRIM25ΔRING-MS2 fusion proteins were co-transfected with the psiCHECK2 constructs in to HEK293 TRIM25 KO cells and processed with dual luciferase assay. (H) Renilla mRNA levels were assayed upon reverse transcription, followed by DNase treatment and qPCR. Renilla mRNA with and without nine MS2 stem loops was measured after mock or T7-TRIM25-MS2 overexpression. (I) Actinomycin D treatment was preformed after co-expressing the psiCHECK2 plasmid containing nine MS2 stem loops with T7-TRIM25-MS2 fusion protein. Renilla mRNA levels were measured upon reverse transcription, followed by DNase treatment and qPCR at 0, 2, 4 and 6 h following actinomycin D treatment. The means and SDs of three (D, E, H, I) or six (F, G) independent experiments are shown. Statistical significance was calculated using one-way ANOVA with Dunnett's (D, E) or Sidak's (H) post-test, two-way ANOVA with Sidak's post-test (I) and ANOVA on ranks (F, G) The asterisk (*) indicates P < 0.05.