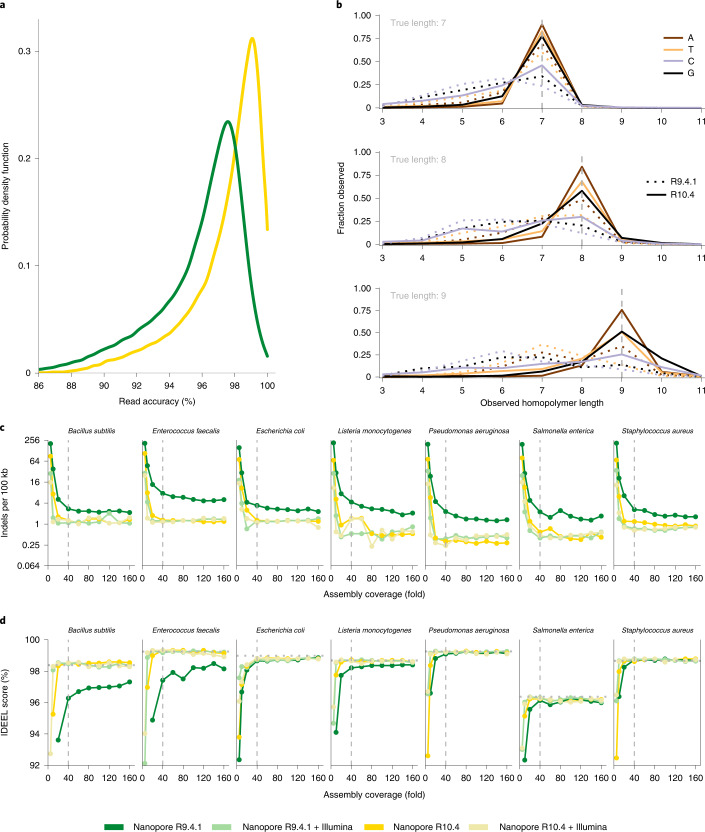
Fig. 1. Sequencing and assembly statistics for the Zymo mock bacterial species (n = 7).
a, Observed raw read accuracies measured through read-mapping. b, Observed homopolymer length of raw reads compared with the reference genomes (see Supplementary Figs. 2 and 3 for a complete overview). c, Observed indels of de novo assemblies per 100 kbp at different coverage levels, with and without Illumina polishing. Note that the reference genomes available for the Zymo mock are not identical to the sequenced strains (Supplementary Table 3). d, IDEEL28 score, calculated as the proportion of predicted proteins that are ≥95% the length of their best-matching known protein in a database19. The dotted line represents the IDEEL score for the reference genome, while the dashed lines mark a 40-fold coverage cut-off.