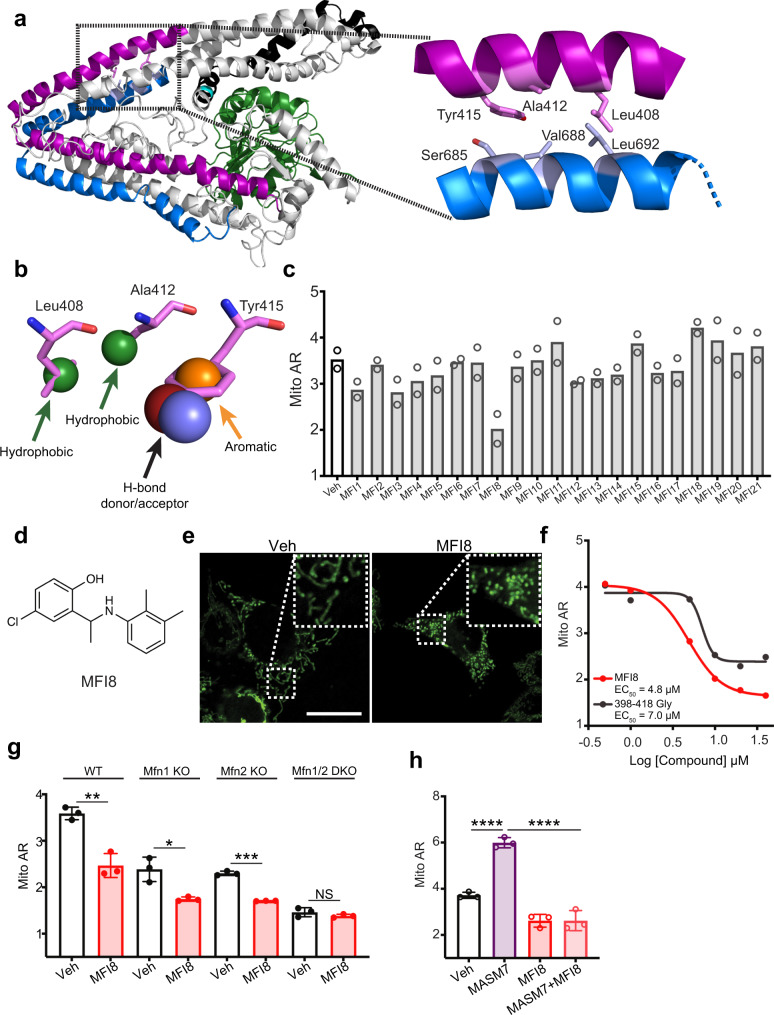
Fig. 2. A small molecule inhibitor of mitofusins.
a Ribbon presentation of the human full-length MFN2 structural model highlighting specific interactions between the HR1 (purple) and HR2 (blue) helical segments. b Pharmacophore hypothesis based on the sidechains of the HR1-amino acids: Leu408, Ala412, Tyr415 interacting with HR2 as in (a) comprising 2 hydrophobic points, one aromatic ring, and one hydrogen bond donor or acceptor. c Screening of putative MFIs using Mito AR as readout. Cells were treated with MFIs (10 μM, 6 h). Data represent mean of two independent biological replicates. d Chemical structure of MFI8. e Confocal micrographs of MEFs treated with MFI8 (20 μM, 6 h). Mitochondria were stained with Mitotracker green. Scale bar 20 μm. Micrograph is representative of n = 3 independent experiments. f MFI8 and 398–418Gly concentration-responsively decreased Mito AR in MEFs. Cells were treated with MFI8 or 398–418Gly at the indicated concentrations for 6 h. Data represent mean of two independent biological replicates. g Quantification of Mito AR of WT, Mfn1 KO, Mfn2 KO, and Mfn1/Mfn2 DKO MEFs treated with MFI8 (20 μM, 6 h). Data represent mean ± SEM of three independent biological replicates. Statistics were obtained using two-tailed unpaired t-test: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. h Quantification of Mito AR of MEFs treated with MASM7 (1 μM), MFI8 (20 μM), and the combination of MASM7 and MFI8 for 6 h. Data represent mean ± SEM of three independent biological replicates. Statistics were obtained using one way ANOVA: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Source data are provided as a Source Data file.