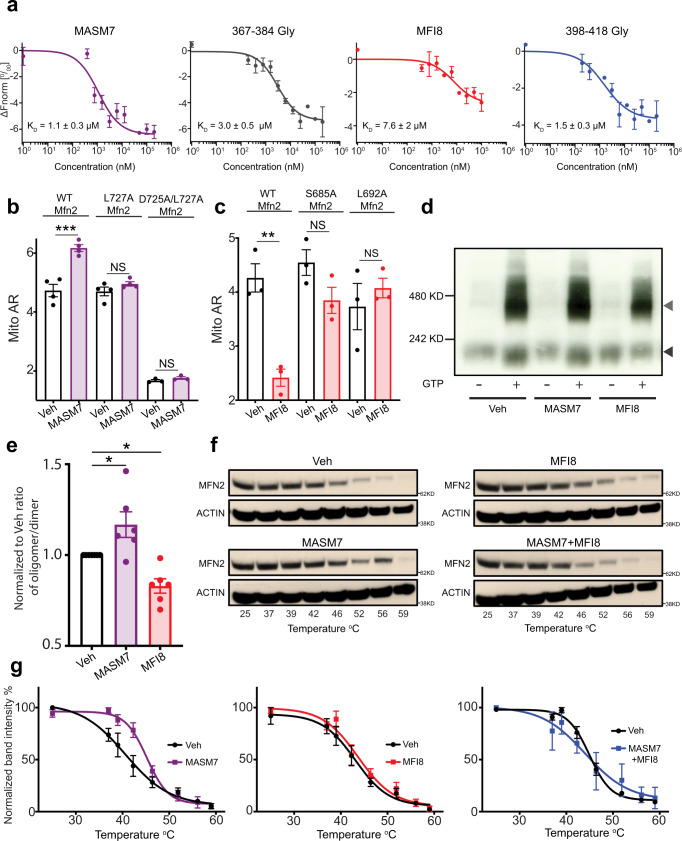
Fig. 3. MASM7 and MFI8 interact specifically with the HR2 domain of MFN2.
a Plots of mean corrected normalized fluorescence (ΔFnorm: Fnomr bound − Fnorm unbound) from MST signal analysis of titrations of HR2 with indicated compounds and peptides. Data represent mean ± SEM from three replicate experiments. b Quantification of Mito AR of Mfn1/Mfn2 DKO MEFs reconstituted with WT, L727A, and D725A/L727A Mfn2 and treated with MASM7 (1 μM, 2 h). Data represent mean ± SEM of three independent biological replicates. Statistics were obtained using two-tailed unpaired t-test: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. c Quantification of Mito AR of Mfn1/Mfn2 DKO MEFs reconstituted with WT, S685A and L692A Mfn2 and treated with MFI8 (20 μM, 6 h). Data represent mean ± SEM of three independent biological replicates. Statistics were obtained using two-tailed unpaired t-test: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. d Separation of MFN2 oligomers from isolated mitochondria treated with GTP, MASM7, and MFI8 as indicated using BN-PAGE electophoresis. Gray arrowhead marks 450 kD MFN2 oligomer and black arrowhead marks MFN2 dimer. e Quantification of the proportion of the MFN2 oligomer observed in the ∼450 kD band to the MFN2 dimer band after treatment with GTP (d). Data represent mean ± SEM of seven independent biological replicates. Statistics were obtained using one way ANOVA: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. f Cellular engagement (CETSA) of MFN2 by MASM7, MFI8, and the combination of both. A representative blot from three independent experiments is shown. g Quantification of temperature-dependent normalized MFN2 levels obtained by densitometry with corresponding fitted curves. Data represent mean ± SEM from n = 3 independent experiments. Source data are provided as a Source Data file.