TABLE 1.
1PA, 2PA and emission properties of NIR-II chromophores reported in literature. Solvent and method of 2P properties are also noted for comparison.
| Probe | Chemical structure | Solvent | λ1PA max | λem max | Φf | 2λ1PA max | λ2PA | σ2 (λ) | σ2 (λ)Φf | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| (nm) | (nm) | (nm) | (nm) | (GM) a | (GM) a | |||||
| Xanthenes | ||||||||||
| Disodium fluorescein (1) |

|
pH11 | 497 | 518 c | 0.90 c | 994 | 1000 | 2.7 | 2.4 b | (Makarov et al., 2008; Mütze et al., 2012) |
| PhenGreen FL (diacetate, uncomplexed) (2) |

|
PBS | 492 c | 517 c | 0.80 c | 984 | 1074 | n.d. | n.d. | (Bestvater et al., 2002) |
| Rhodamine 6G (3) |

|
MeOH | 519 c | 546 c | 0.95 c | 1038 | 1060 | 10 | 9.5 b | (Makarov et al., 2008) |
| Rhodamine B (4) |

|
MeOH | 553 c | 627 c | 0.70 c | 1106 | 1040 | 39 | 27 b | Makarov et al., 2008 |
| Rhodamine 101 (5) |
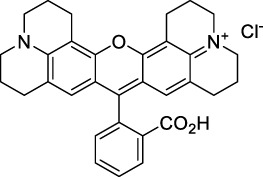
|
EtOH | 570 c | 591 c | 1.0 c | 1140 | 1060 | 20 | 20 b | (Li and She, 2010; Mütze et al., 2012) |
| Rhodamine 123 (6) |

|
PBS | 507 | 529 c | 0.90 c (EtOH) | 1014 | 1090 | n.d. | n.d. | (Bestvater et al., 2002) |
| Alexa Fluor 488 (7) |

|
NaPhos | 491 | 519 c | 0.92 c | 980 | 1000 | 21 b | 19 | (Bestvater et al., 2002; Anderson and Webb, 2011; Mütze et al., 2012) |
| Alexa Fluor 546 (8) |

|
PBS | 553 | 573 c | 0.79 c | 1112 | 1028 | n.d. | n.d. | (Bestvater et al., 2002; Mütze et al., 2012) |
| Alexa Fluor 568 (9) |

|
PBS | 578 c | 603 c | 0.69 c | 1156 | 1060 | n.d. | n.d. | (Mütze et al., 2012) |
| Alexa Fluor 594 (10) |

|
PBS | 594 c | 617 c | 0.66 c | 1180 | 1074 | n.d. | n.d. | (Bestvater et al., 2002; Mütze et al., 2012) |
| Alexa Fluor 610 (11) |

|
PBS | 612 c | 628 c | – | 1224 | 1010 | n.d. | n.d. | (Mütze et al., 2012) |
| Alexa Fluor 633 (12) |

|
H2O | 632 c | 647 c | – | 1264 | 1260 | n.d. | <5 | (Kobat et al., 2009; Mütze et al., 2012) |
| MitoTracker Red (13) |

|
PBS | 579 c | 599 c | 0.15 [187] | 1158 | 1133 | n.d. | n.d. | (Bestvater et al., 2002) |
| CellTracker Red (14) |

|
In vitro | 585 c | 602 c | n.d. | 1170 | 1080 | n.d. | n.d. | (Rakhymzhan et al., 2017) |
| Lissamine Rhodamine-IgG (15) |

|
PBS | 570 c | 590 c | 0.33 [188] | 1140 | 1116 | n.d. | n.d. | (Bestvater et al., 2002) |
| Texas Red-IgG (16) |

|
PBS | 596 c | 615 c | 0.90 c | 1192 | 1150 | n.d. | n.d. | (Bestvater et al., 2002) |
| ATTO 680 (17) |

|
In vitro | 681 c | 698 c | 0.30 c | 1362 | 1260 | n.d. | n.d. | (Rakhymzhan et al., 2017) |
| Nile Red (18) |

|
MeOH | 550 | 636 | 0.40 | 1100 | 1057 | 104 | 42 | (Hornum et al., 2020) |
| 19 |

|
MeOH | 554 | 631 | 0.43 | 1108 | 1055 | 183 | 79 | (Hornum et al., 2020) |
| 20 |

|
MeOH | 569 | 632 | 0.45 | 1065 | 1050 | 123 | 55 | (Hornum et al., 2020) |
| 21 |

|
MeOH | 565 | 638 | 0.35 | 1130 | 1057 | 232 | 81 | (Hornum et al., 2020) |
| Polymethines | ||||||||||
| Cy3-IgG (22) |

|
PBS | 548 c | 563 c | 0.1 c | 1096 | 1032 | n.d. | n.d. | (Bestvater et al., 2002) |
| Cy5 (23) |

|
H2O | 646 c | 662 c | 0.28 c | 1292 | 1220 | 143 b | ≈40 | (Kobat et al., 2009) |
| Cy5.5 (24) |

|
H2O | 673 c | 691 c | 0.21 c | 1346 | 1280 | 286 b | ≈60 | (Kobat et al., 2009) |
| Cy7 (25) |

|
H2O | 750 c | 773 c | 0.30 c | 1500 | 1320 | 200 b | ≈60 | (Kobat et al., 2009) |
| 26 |

|
DMSO | 753 | 780 | 0.17 | 1506 | 1552 | 240 | 41 b | (Berezin et al., 2011) |
| ICG (27) |

|
DMSO | 794 | 817 | 0.12 | 1588 | 1552 | 590 | 71 b | (Berezin et al., 2011) |
| Cypate (28) |

|
DMSO | 796 | 817 | 0.13 | 1592 | 1552 | 520 | 68 b | (Berezin et al., 2011) |
| 29 |

|
DMSO | 809 | 829 | 0.07 | 1618 | 1552 | 900 | 63 b | (Berezin et al., 2011) |
| DTTC (30) |

|
DMSO | 771 c | 800 c | 0.80 c | 1542 | 1552 | 160 | 128 b | (Berezin et al., 2011) |
| DODCI (31) |
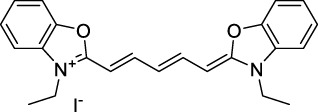
|
EtOH | 582 c | 610 c | 0.87 c (DMSO) | 1164 | 1060 | 38 | n.d. | (Li and She, 2010) |
| IR-140 (32) |

|
DMSO | 825 | ≈840 | 0.06 | 1640 | 1552 | 950 | 57 b | (Berezin et al., 2011) |
| 33 |

|
CH2Cl2 | 1064 | ≈1080 | 0.05 | 2128 | 1800 | 2250 | 113 b | (Hu et al., 2013) |
| CH3CN | 1043 | ≈1065 | 0.05 | 2086 | 1800 | 1050 | 53 b | |||
| 34 |

|
EtOH | 650 | 665 | n.d. | 1300 | 1180 | 140 | n.d. | (Fu et al., 2007) |
| 35 |

|
EtOH | 690 | 704 | n.d. | 1380 | 1260 | 150 | n.d. | (Fu et al., 2007) |
| 36 |

|
EtOH | 770 | n.d. | n.d. | 1540 | 1340 | 60 | n.d. | (Fu et al., 2007) |
| 37 |

|
EtOH | 824 | n.d. | n.d. | 1648 | 1480 | 600 | n.d. | (Fu et al., 2007) |
| Alexa Fluor 647 (38) |

|
H2O | 650 c | 665 c | 0.33 c | 1300 | 1240 | 133 b | ≈44 | (Kobat et al., 2009; Mütze et al., 2012) |
| Alexa Fluor 680 (39) |

|
H2O | 679 c | 702 c | 0.36 c | 1358 | 1280 | 203 b | ≈73 | (Kobat et al., 2009) |
| Alexa Fluor 700 (40) | – d | H2O | 702 c | 723 c | 0.25 c | 1404 | 1320 | 208 b | ≈52 | (Kobat et al., 2009) |
| Alexa Fluor 750 (41) | – d | H2O | 753 c | 778 c | 0.12 c | 1506 | 1320 | 292 b | ≈35 | (Kobat et al., 2009) |
| 42 |

|
MeOH | 532 | 636 | 0.44 | 1064 | 1064 (900) | 23 (570) | 10 b | (Pascal et al., 2017) |
| 43 |

|
MeOH | 573 | 708 | 0.33 | 1146 | 1146 | 225 | 74 b | (Pascal et al., 2017) |
| 44 |

|
MeOH | 549 | 673 | 0.54 | 1098 | 1098 | 137 | 74 b | (Pascal et al., 2017) |
| 45 |

|
Toluene | 643 | 654 | 0.62 | 1286 | 1198 | 133 | 82 b | (Ceymann et al., 2016) |
| 46 |

|
Toluene | 700 | 714 | 0.75 | 1400 | 1274 | 100 | 75 b | (Ceymann et al., 2016) |
| Styryl 9M (47) |

|
CHCl3 | ≈625 | ≈790 | 0.10 [189] | ≈1250 | 1240 | 780 | 78 | (Makarov et al., 2008) |
| FM4-64 (48) |

|
PBS CHCl3 (López-Duarte et al., 2015) |
471 564 |
691 761 |
n.d. 0.35 |
942 1128 |
1047 | n.d. | n.d. | (Wokosin et al., 1996a; Nuriya et al., 2016) |
| TO-PRO-3 (49) |

|
H2O | 641 c | 657 c | n.d. | 1284 | 1110 | n.d. | n.d. | (Smith et al., 2012) |
| 50 |

|
THF | 562 | 598 | 0.07 | 1124 | 1070 | 167 | 12 b | (Poronik et al., 2012) |
| 51 |

|
THF | 615 | 655 | 0.02 | 1230 | 1150 | 214 | 4 b | (Poronik et al., 2012) |
| Porphyrins | ||||||||||
| 52 |

|
CCl4 | ≈770 | ≈780 | n.d. | ≈1540 | 1020 1270 |
470 48 |
n.d. | (Makarov et al., 2008) |
| 53 |

|
CCl4 | ≈685 | ≈700 | n.d. | ≈1370 | 1270 | 13 | n.d. | (Makarov et al., 2008) |
| 54 |

|
CHCl3 | ≈525 ≈605 ≈680 |
710 | n.d. | ≈1050 ≈1210 ≈1360 |
≈1040 ≈1220 ≈1360 |
≈2000 ≈500 ≈200 |
n.d. | (Nowak-Król et al., 2013) |
| BODIPYs | ||||||||||
| LysoTracker Red (55) |

|
PBS | 577 c | 590 c | 0.07 | 1154 | 1100 | n.d. | n.d. | (Bestvater et al., 2002) |
| BODIPY-TR (56) |

|
MOPS | 589 c | 616 c | 0.90 c | 1178 | 1060 | 269 b | 242 | (Bestvater et al., 2002; Mütze et al., 2012) |
| IR-07 (57) |

|
CH2Cl2 | ∼700 | 750 | 0.30 | ∼1400 | 1310 | 101 | 30 b | (Zheng et al., 2009) |
| 58 |

|
THF | 755 | 830 | 0.09 | 1560 | 1064 | n.d | n.d | (Hu et al., 2020) |
| Dipoles – Quadrupoles – Miscellaneous | ||||||||||
| 59 |

|
NPs (Aq.) | 480 | 678 | 0.17 | 960 | 1040 | 5.6 × 105 | 9520 | (Alifu et al., 2017) |
| 60 |

|
H2O (0.1% DMSO) | 530 | 740 | n.d. | 1060 | 1100 | n.d. | n.d. | (Zhou et al., 2021) |
| 61 |

|
H2O | 510 | 676 | 0.22 | 1020 | 1040 | 440 | 97 b | (Massin et al., 2013) |
| 62 |

|
CH2Cl2 | 660 | 785 | 0.005 | 1320 | 1300 | 500 | 2.5 b | (Ricci et al., 2017) |
| 63 |

|
CH2Cl2 | 678 | 782 | 0.0005 | 1356 | 1300 | 1400 | 0.7 b | (Ricci et al., 2017) |
| 64 |

|
H2O | ≈600 | ≈725 | 0.21 | ≈1200 | 1200 | 1.21 × 103 | 242 b | (Wang et al., 2019b) |
| 65 |

|
CHCl3 | 634 | 704 | 0.16 | 1268 | 1250 | 920 | 147 b | (Li et al., 2012) |
| 66 |

|
CHCl3 | 668 | 807 | 0.02 | 1336 | 1250 | 1200 | 24 b | (Li et al., 2012) |
| 67 |

|
CHCl3 | 1088 | 1120 | 0.002 | 2176 | 2200 | 1300 | 2.6 b | (Ni et al., 2016) |
| 68 |

|
CHCl3 | 1136 | 1193 | 0.0002 | 2272 | 2300 | 1500 | 0.3 b | (Ni et al., 2016) |
| Propidium iodide (69) |

|
PBS | 536 c | 617 c | 0.20 c (dsDNA bound) | 1072 | 1015 | n.d. | n.d. | (Bestvater et al., 2002) |
| AIEgens and AIEDots | ||||||||||
| 70 |

|
NP (aq.) | 613 | 790-810 | 0.14 | 1226 | 1040 1300 |
16100 1220 |
2240
b
170 b |
(Qi et al., 2018; Liu et al., 2021) |
| 71 |

|
NP (aq.) THF |
454 ≈451 |
≈700 ≈699 |
0.19 n.d. | 908 | 1200 n.d. | 76300 n.d. | 14500 b n.d. | (Wang et al., 2019c) |
| 72 |

|
NP (aq.) H2O/DMSO |
≈479 ≈488 |
≈627 >627 |
0.06 n.d. | ≈960 | 1040 n.d. | 3200 n.d. | 192 b n.d. | (Samanta et al., 2021) |
| 73 |

|
NP (aq.) Toluene |
510 491 |
709 ≈635 |
0.14 (solid state) | 1020 | 1000 n.d. | ≈520 n.d. | 73 b n.d. | (Zheng et al., 2018) |
| 74 |

|
NP (aq.) Toluene |
538 528 |
755 ≈636 |
0.02 (solid state) | 1076 | 1020 n.d. | 887 n.d. | 18 b n.d. | (Zheng et al., 2018) |
| 75 |

|
NP (aq.) THF |
522 511 |
620 532 |
0.05n.d. | 1044 | 1040 n.d. | 2.9 ×106n.d. | 1.5 × 105n.d. | (Wang et al., 2015) |
| Carbon, hybrid and inorganic materials | ||||||||||
| 76 | SWCNT-based dopamine sensor | H2O | 600–1000 | 1000–1265 e | 0.0023 | – | 1560 | 216000 | 497 b | (Bonis-O'Donnell et al., 2017) |
| 77 | Aptamer-modified graphene oxide | H2O | 440–720 | 500–650 f | 0.34 | – | 1120 | 36000 | 12240 b | (Pramanik et al., 2014) |
| 78 | CDs prepared from urea and citric acid | H2O | 540 | 624 | 0.06 | 1080 | 1200 | n.d. | n.d. | (Li et al., 2018) |
| 79 | Carbon quantum dots prepared from tris(4-aminophenyl)amine | H2O | 592 | 615 | 0.84 | 1184 | 1100 | n.d. | n.d. | (Liu et al., 2020) |
| 80 | AuNP with SWCNT | H2O | 500–1100 | 775 | n.d. | – | 1100 | n.d. | n.d. | (Olesiak-Banska et al., 2019) |
| 81 | Au25 cluster | H2O | 675 | 830 | <0.001 | 1350 | 1290 | 2700 | n.d. | (Ramakrishna et al., 2008) |
| 82 | PEG-dithiolane AuNC | H2O | 355, 670 | 820 | 0.08 | 1370 | 1100 | 300 | 24 b | (Oh et al., 2013) |
| 83 | Mn2+-ZnS QD | H2O | 318 | 586 | 0.65 | 636 | 1180 | 265 | 172 b | (Subha et al., 2013) |
| 84 | PbS/CdS QD | H2O | 665 | 1270 | 0.18 | 1330 | 1550 | 530 | 95 b | (Ni et al., 2022) |
| QD605 (85) | polymer-encapsulated CdSe-ZnS QD | H2O | 350–475 | 605 | 0.71 | – | 1000 | 66200 b | 47000 | (Larson et al., 2003) |
| Fluorescent proteins | ||||||||||
| tdTomato (86) | – | H2O | 554 g | 581 | 0.72 b | 1108 | 1050 | 278 | 200 | (Drobizhev et al. 2011) |
| tdKatushka2 (87) | – | H2O | 588 g | 633 | 0.44 b | 1176 | 1100 | 143 | 63 | (Drobizhev et al. 2011) |
| dsRed2 (88) | – | H2O | 561 g | 587 | 0.71 b | 1126 | 1050 | 103 | 73 | (Drobizhev et al. 2011) |
| HcRFP (89) | – | PBS | 592 g | 645 g | 0.05 g | 1184 | 1160 | 720 b | 36 | (Tsai et al., 2006) |
| mCherry (90) | – | H2O | 587 g | 610 | 0.24 b | 1174 | 1080 | 27 | 6.4 | (Drobizhev et al. 2011) |
| mBanana (91) | – | H2O | 540 g | 553 | 0.69 b | 1080 | 1070 | 64 | 44 | (Drobizhev et al. 2011) |
| mStrawberry (92) | – | H2O | 574 g | 596 | 0.34 b | 1148 | 1070 | 20 | 6.8 | (Drobizhev et al. 2011) |
| mRFP (93) | – | H2O | 584 g | 611 | 0.30 b | 1168 | 1080 | 44 | 13 | (Drobizhev et al. 2011) |
| TagRFP (94) | – | H2O | 555 g | 584 | 0.44 b | 1110 | 1050 | 95 | 42 | (Drobizhev et al. 2011) |
| mOrange (95) | – | H2O | 548 g | 565 | 0.70 b | 1096 | 1080 | 67 | 47 | (Drobizhev et al. 2011) |
| eqFP650 (96) | – | H2O | 592 g | 646 | 0.19 b | 1184 | 1112 | 45 | 8.5 | (Drobizhev et al. 2011) |
| Katushka (97) | – | H2O | 588 g | 635 | 0.35 b | 1176 | 1080 | 66 | 23 | (Drobizhev et al. 2011) |
| Katushka2(98) | – | H2O | 588 g | 633 | 0.44 b | 1176 | 1140 | 62 | 27 | (Drobizhev et al. 2011) |
| mKate (99) | – | pH8 | 588 g | 635 | 0.27 b | 1176 | 1118 | 52 | 14 | (Drobizhev et al. 2011) |
| mKate2 (100) | – | H2O | 588 g | 633 | 0.42 b | 1176 | 1140 | 72 | 30 | (Drobizhev et al. 2011) |
| mNeptune (101) | – | H2O | 600 g | 651 | 0.17 b | 1200 | 1104 | 70 | 12 | (Drobizhev et al. 2011) |
| mRaspberry (102) | – | H2O | 598 g | 625 | 0.19 b | 1196 | 1118 | 31 | 5.8 | (Drobizhev et al. 2011) |
| Neptune (103) | – | H2O | 600 g | 647 | 0.22 b | 1200 | 1104 | 72 | 16 | (Drobizhev et al. 2011) |
| tdRFP (104) | – | Aq. buffer | 584 (Campbell et al., 2002) | 579 | 0.68 | 1168 | 1110 | 20 | 13.7 | (Drobizhev et al. 2011) |
Two-photon absorption cross-section value taken at the excitation wavelength λ2PA reported by the authors in the NIR-II window; note that this may differ from the maximum of the 2PA band. Value extrapolated from the data available and from the formula of the 2P brightness (= σ2 (λ) × ΦF).
1P properties as reported by commercial suppliers, reported in water unless indicated otherwise.
Chemical structures are propriety and undisclosed.
Chirality-dependant.
Excitation-dependant.
Properties extracted from the fluorescent protein database (Available at https://www.fpbase.org/, Accessed on 11/04/2022).
