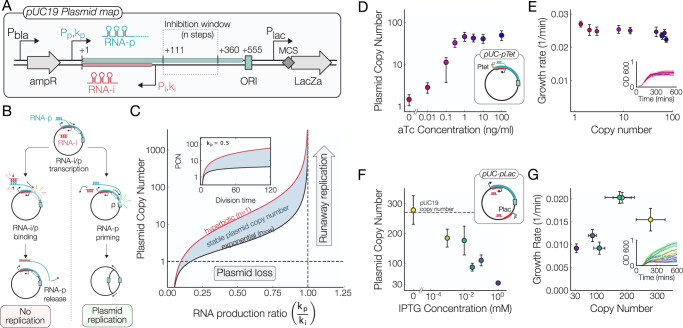
Fig. 1. Replication of ColE1 origin plasmids.
A Map of the pUC19 plasmid. The origin of replication consists of a 555 bp region containing the priming RNA (RNA-p), the inhibitory RNA (RNA-i), the inhibition window, and the site of replication initiation (ORI). B Diagram of the steps leading to ColE1 replication. In the ColE1 Origin, reversible binding of the inhibitory RNA-i to RNA-p exercises control over the copy number by inhibiting replication (left branch). Production of RNA-p transcript triggers plasmid replication through the hybridization of RNA-p with the ORI (right branch). C Predicted plasmid copy number for hyperbolic (upper) and exponential (lower) initiation models28 plotted as a function of RNA-p transcription initiation rate. Inset shows the predicted dependence of plasmid copy number on host cell division time. Model parameters: ϵ = 0.545 min, ρ = 0.65, r = 0.01 min, ki = 0.95 min. D Plasmid copy number measurements made by digital droplet PCR of the aTc-inducible pUC-pTet plasmid at increasing aTc concentrations; n = 3, error bars derived from Poisson distribution of ddPCR counts. E Growth rates of cells hosting the pUC-pTet plasmid at increasing plasmid copy numbers. We see a marginal effect of plasmid copy number on cellular growth rates, inset shows individual growth curves colored by aTc concentration. Data points are means across n = 6 biological replicates, error bars = std. dev. F Plasmid copy number measurements of a plasmid that contains an additional copy of the inhibitory RNA under the control of an IPTG-inducible promoter; n = 3, error bars derived from Poisson distribution of ddPCR counts. G Growth rate vs plasmid copy number of the IPTG-inducible plasmid shows a strong metabolic burden at high levels of RNA-i expression. Data points are means across n = 6 biological replicates, error bars = std. dev.