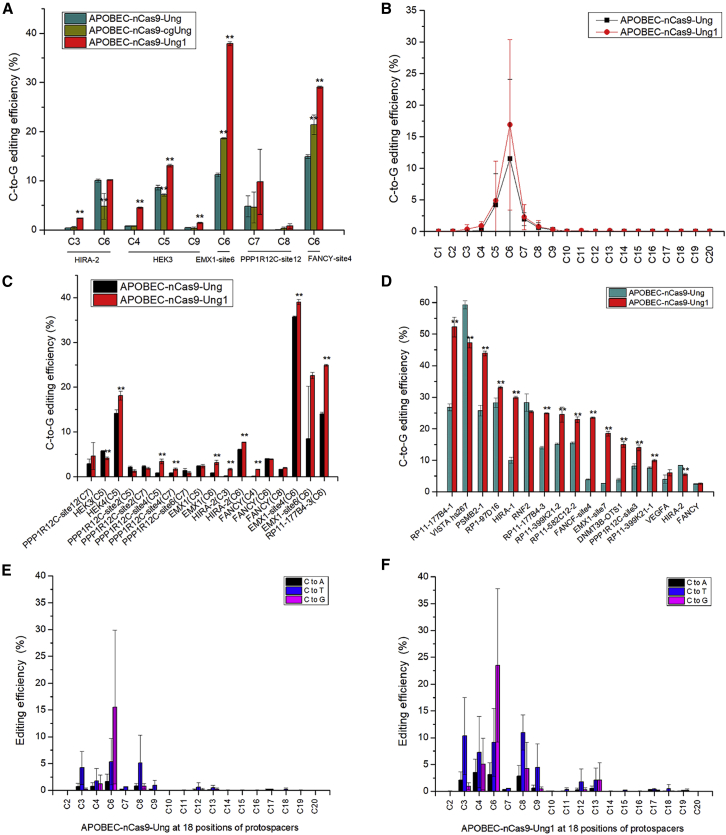
Figure 1.
Base editing with different GBEs in HEK293T cells
(A) The C-to-G base editing efficiency of GBEs with different Ung homologs. (B) The average C-to-G base editing efficiency of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 at 20 positions of protospacers from 12 loci (6 gRNA for non-C6 sites and 6 for C6 sites). (C) The C-to-G base editing efficiency of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 at the C3–C8 position from 12 loci (6 gRNA for non-C6 sites and 6 for C6 sites). (D) The C-to-G base editing efficiency of APOBEC-nCas9-Ung and APOBEC-nCas9-Ung1 at the C6 position of 17 loci. (E) The average base editing (C-to-A, C-to-T, and C-to-G) efficiency of APOBEC-nCas9-Ung at 18 positions of protospacers from 17 loci (gRNA for C6 sites). (F) The average base editing (C-to-A, C-to-T, and C-to-G) efficiency of APOBEC-nCas9-Ung1 at 18 positions of protospacers from 17 loci (gRNA for C6 sites). ∗∗p < 0.01 was considered statistically significant. The bars represent the average editing efficiency, and error bars represent the SD of 3 independent biological replicates.