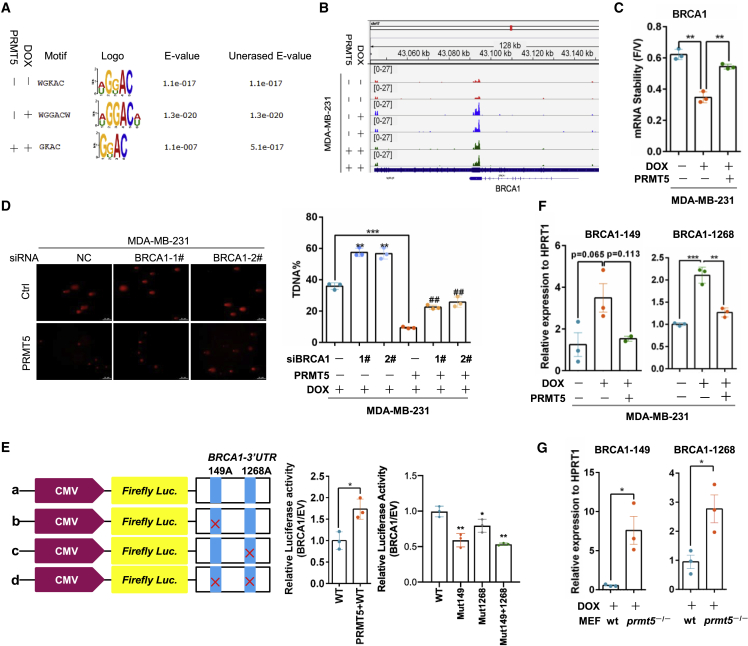
Figure 2.
PRMT5 decreases the m6A methylation of BRCA1 mRNA to increase BRCA1 stability
(A) The m6A peak enrichment was analyzed in each group as indicated and characterized by the canonical GGAC motif. (B) The m6A abundances in BRCA1 mRNA transcripts in different groups were detected by m6A-seq. (C) MDA-MB-231 cells were treated with 0.5 μg/mL DOX for 24 h and then treated with vehicle or 3.2 μM flavopiridol for 6 h. BRCA1 mRNA levels were measured by RT-qPCR, and the flavopiridol/vehicle (F/V) ratio of BRCA1 mRNA was determined (mean ± SEM; n = 3). (D) A comet assay was performed with MDA-MB-231 cells with or without PRMT5 overexpression and BRCA1-specific siRNA treated with 0.4 μg/mL DOX for 24 h. Right: quantification of the comet tail that represents broken DNA strands (T-DNA%). ##p < 0.01 versus MDA-MB-231-PRMT5-siNC, ∗∗p < 0.01 versus MDA-MB-231-siNC, and ∗∗∗p < 0.001 versus MDA-MB-231-siNC. NC, negative control. (E) A relative luciferase assay was performed to analyze the BRCA1 mRNA 3′-UTR with intact or mutant m6A sites. (F) RT-PCR analysis was performed following m6A-IP of the BRCA1 mRNA 3′-UTR in MDA-MB-231 cells with PRMT5 overexpression treated with 0.4 μg/mL doxorubicin for 24 h. BRCA1-149 and BRCA1-1268 represent two different m6A sites in the BRCA1 mRNA 3′-UTR. ∗∗p < 0.01, and ∗∗∗p < 0.001. (G) RT-PCR analysis was performed following m6A-IP of the BRCA1 3′-UTR in wild-type and prmt5-knockout MEFs treated with 0.4 μg/mL doxorubicin. ∗p < 0.05.