Figure 4.
FOSL1 induces activation of NF-κB signaling
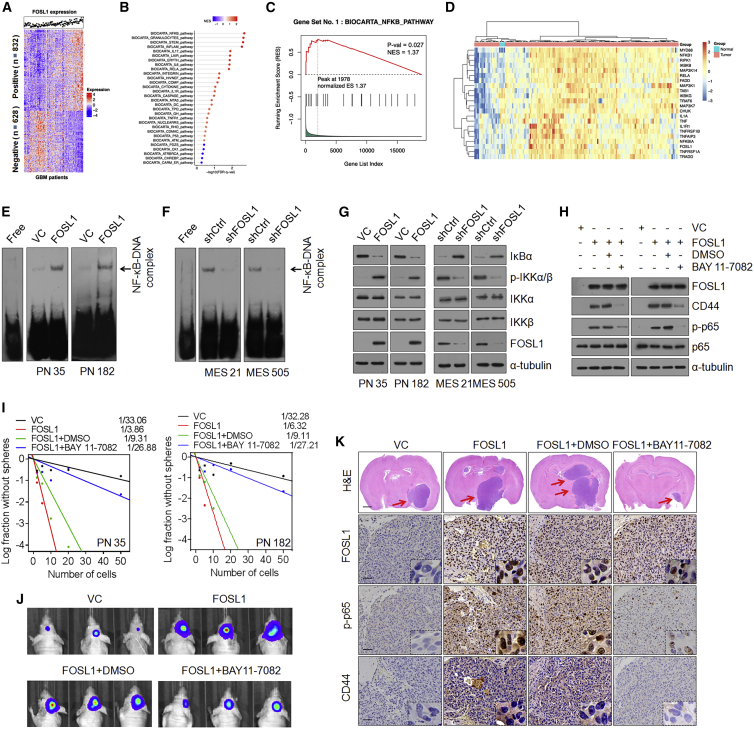
(A) Cluster heatmap showing 832 FOSL1 positively correlated and 628 FOSL1 negatively correlated genes in TCGA database. (B) Gene set enrichment analysis (GSEA) showing signaling pathways related to FOSL1 in human primary GBM. The color gradation and location of the dot respectively indicate the GSEA score and p value. (C) Correlation between the enrichment of FOSL1 and NF-κB pathway gene expression in GBM by GSEA analysis. (D) Heatmap depicting genes differentially expressed between FOSL1 and NF-κB pathways/targets in normal brain tissues (n = 5) and GBM (n = 168). Genes are labeled and clustered using hierarchical clustering. (E and F) EMSA analysis of NF-κB DNA-binding activity in PN 35 and PN 182 (E), and MES 21 and MES 505 (F) GSCs with indicated modifications. (G) IB analysis of IκBα, p-IKKα/β (Ser180/181), IKKα, IKKβ, and FOSL1 in PN 35, PN 182, MES 21, and MES 505 GSCs with indicated modifications. α-Tubulin was used as internal control. (H) IB analysis of FOSL1, CD44, C/EBPβ, TAZ, p-STAT3 (Tyr705), STAT3, p-p65 (Ser536), and p65 in PN 35 and PN 182 GSCs transduced with vector control or FOSL1 in the presence or absence of BAY 11-7082. α-Tubulin was used as internal control. (I) Limiting dilution neurosphere-forming assay in PN 35 and PN 182 GSCs expressing FOSL1 or vector control, treated with dimethyl sulfoxide (DMSO) or BAY 11-7082. (J) Representative BLI images of mice bearing xenografts derived from luciferase-expressing PN 35 and PN 182 GSCs transduced with FOSL1 or vector control, with or without BAY 11-7082 treatment. (K) Representative H&E-stained brain sections and IHC-staining images of FOSL1, p-p65, and CD44 in indicated PN GSC-derived xenografts treated with DMSO or BAY 11-7082. Red arrows indicate tumors. Scale bars, 1 mm (H&E staining) and 25 μm (IHC staining).