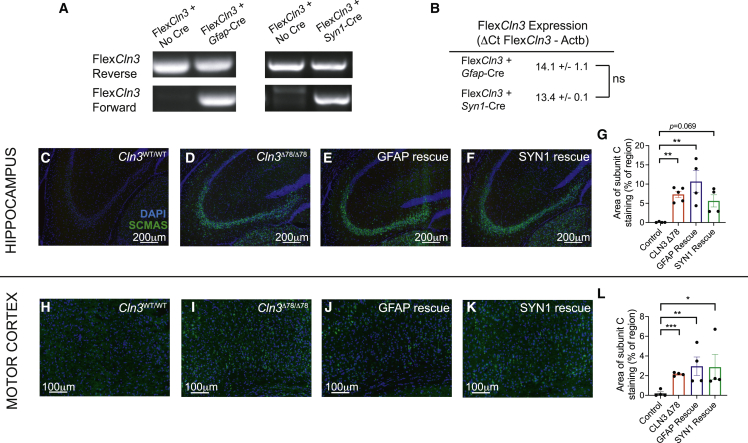
Figure 3.
Expression of FlexCln3 allele in either neurons or astrocytes alone does not prevent storage accumulation in 12-month-old animals
(A) In the presence of Cre, the FlexCln3 allele undergoes inversion as detected by PCR with orientation-specific primers of genomic DNA from hippocampus. As both the Gfap-Cre and Syn1-Cre only drive recombination in a subset of cells, both the forward and reverse alleles are detected (see Figure S7 for uncut gels). (B) Expression levels as measured by ΔCt values (FlexCln3-βactin) are similar in FlexCln3 brain in the presence of Gfap-Cre and Syn1-Cre (14.1 versus 13.4, p = 0.53 by unpaired t test). (C–G) Expression of FlexCln3 in astrocytes (FlexCln3/Cln3Δex78/Δex78/Gfap-Cre animals) or neurons alone (FlexCln3/Cln3Δex78/Δex78/Syn1-Cre animals) does not prevent accumulation of SCMAS (green). (H–L) Similar findings were seen in the motor cortex. N = 4–5 animals, non-parametric Kruskal-Wallis test followed by Benjamini and Hochberg multiple comparisons correction to detect differences from control. For all panels: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.