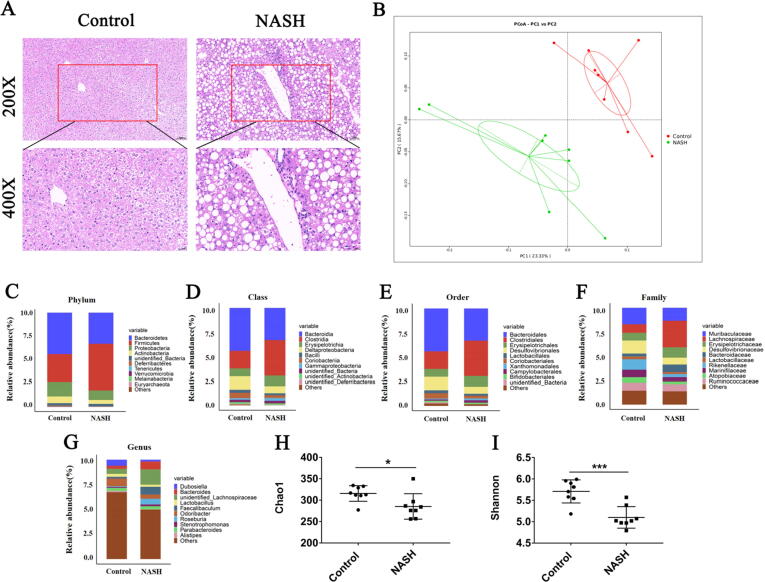
Fig. 1.
Comparison of gut microbiota between control and NASH mice. (A): H&E staining for liver tissues (scale bar: 50 μM for the top and 20 μM for the bottom row); (B): Unweighted UniFrac principal coordinate analysis; (C–G): Most abundant taxa at the phylum (C), class (D), order (E), family (F), and genus (G) levels; (H–I): Chao1 (H) and Shannon (I) alpha diversity metrics were reduced in NASH mice. Data are mean ± SD, n = 8; *p < 0.05, **p < 0.01, ***p < 0.001 versus control group.