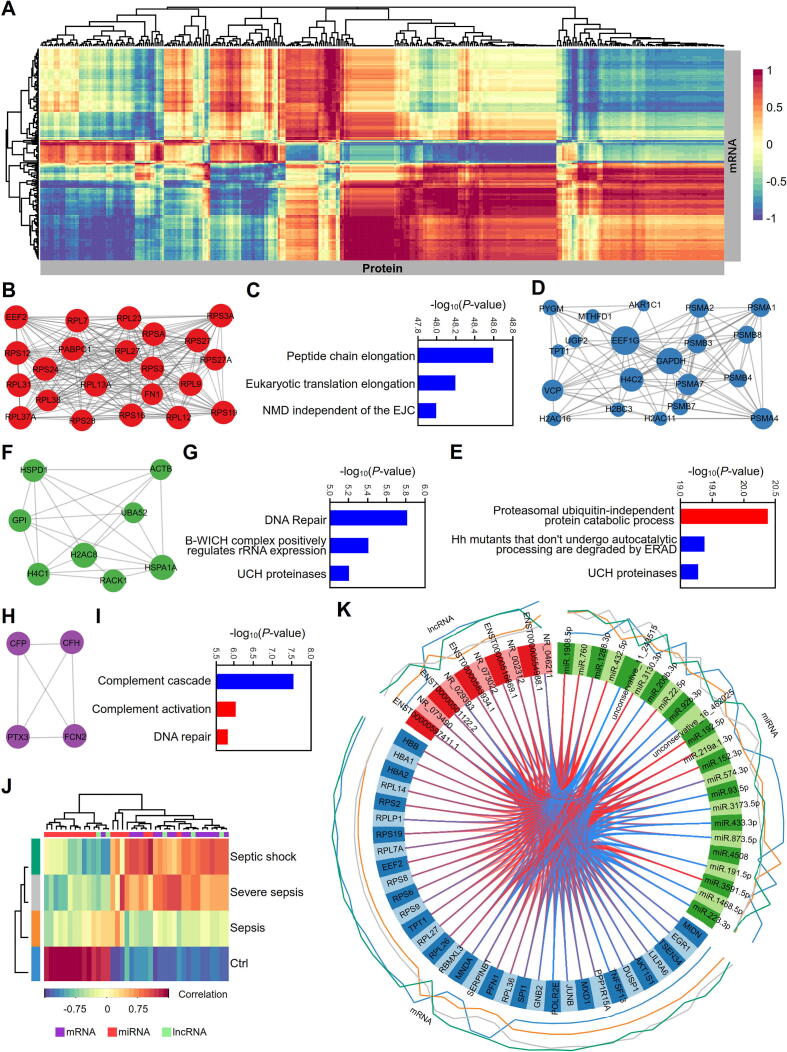
Fig. 8.
Integrative multiomics analysis of proteins, mRNAs, lncRNAs and miRNAs in the serum exosomes of septic patients. (A) Hierarchical clustering for canonical correlation analysis (CCA) of the mRNAs and proteins in serum exosomes from septic patients. The mixOmics R package was used to calculate the correlations between mRNA abundance (rows) and protein abundance (columns) in a pairwise fashion. (B-I) Identification and functional annotation of MCODE subclasses in the interaction networks for the hierarchical cluster described above. The interaction networks were created with the BioGrid, InWeb_IM and OmniPath databases. MCODE analysis was performed to identify densely connected components in the networks (B, D, F, H). Pathway and biological process enrichment analyses were applied to each MCODE component independently, and the three best-scoring terms by P-value were retained as the functional descriptions of the corresponding subclasses (C, E, G, I). The blue color represents the pathway enrichment analysis terms, and the red color represents the biological process terms from GO enrichment analysis. (J) CIM of the multiomics signatures for mRNAs, lncRNAs and miRNAs in the serum exosomes of septic patients. The map was constructed on Euclidian distance and complete linkage with the mixOmics R package for transcriptomic datasets including mRNAs, lncRNAs and miRNAs in serum exosomes from patients with sepsis at different stages. Grouped samples are represented in rows, and selected features on the first component are represented in columns. (K) Circos plot representing the correlations among mRNAs, lncRNAs and miRNAs in serum exosomes from patients with sepsis at different stages. The Circos plot was obtained by using the function circosPlot() from the package mixOmics and shows the positive (red line) and negative (blue line) correlations (|r| >0.7) between variables (edges inside the circle) and the average value of each variable (line profile outside the circle).