Figure 5.
Integrated clustering of transcriptomics and metabolomics using similarity network fusion (SNF) shows novel stratifications for patients based on molecular data
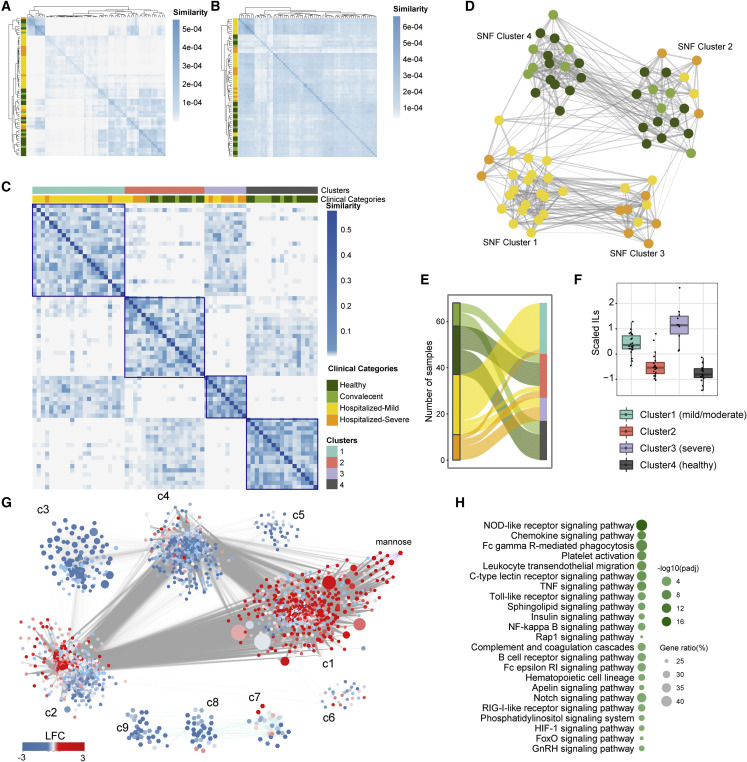
(A) Heatmap showing sample to sample similarity between all samples in each patient cohorts (HC [n = 21], convalescent [n = 10], hospitalized-mild [n = 26], and hospitalized-severe [n = 11]) for transcriptomics data. Row annotation denotes patient cohort.
(B) Heatmap showing sample to sample similarity between all samples used in (A) for metabolomics data. Row annotation denotes patient cohort.
(C) Sample to sample similarity of SNF-derived patient clusters. Column annotation denotes SNF clusters and associated clinical categories.
(D) Network fusion diagram of the four integrated patient clusters. Node color indicates the clinical category of the samples; edges indicate a similarity >0.01. (HC [dark green], convalescent [light green], hospitalized-mild [yellow], and hospitalized-severe [orange])
(E) Sankey plot depicting association of clinical categories, and new clusters defined based on molecular data for each sample.
(F) Mean-scaled interleukins (IL) of plasma inflammation profile identified by the IL6, IL7, IL8, IL10, IL15, and IL18 to define the severity of the data-driven clusters.
(G) Community characterization results obtained from topology analysis of the association network between genes and metabolites among samples of SNF-1, 3, and 4. Node size is relative to the centrality (degree) of the feature (gene/metabolite). Node colors denote log2-fold change values of the feature in SNF-3 (severe, n = 10) compared with SNF-1 (mild/moderate, n = 22). Each edge denotes a significant Spearman correlation (adj. p < 0.00001).
(H) Significantly enriched pathways (adj. p < 0.01) in the most central community found in (F) (community 1) based on average node centrality (degree).