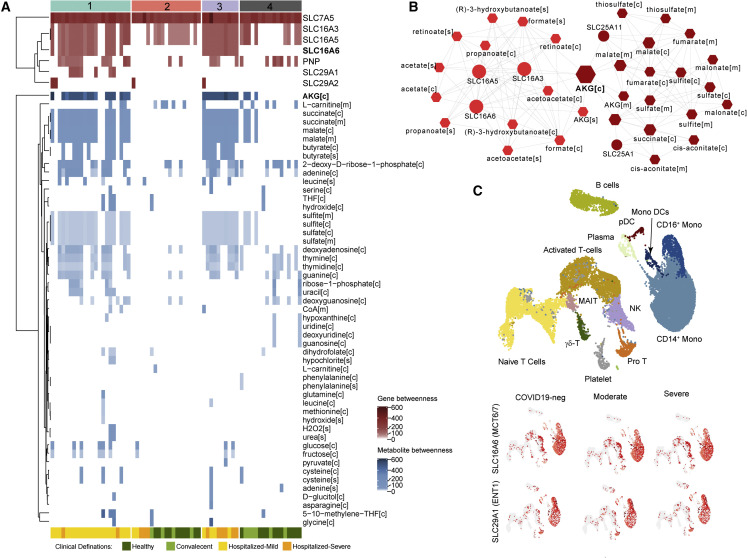
Figure 7.
Network-based essential gene and metabolite analysis reveal the role of transporter genes and TCA-cycle intermediates in COVID-19
(A) The essential genes and metabolites are identified based on topology analysis of the network created using metabolites and associated genes of selected reactions in each sample. The heatmap visualizes the centrality measurements (betweenness) of genes and metabolites in each sample network. Top column annotation denotes sample classification by SNF clustering. The bottom column annotation denotes original cohorts.
(B) Topology analysis results of the network created using metabolites and associated genes of selected reactions in SNF cluster 3 (severe, n = 10). The figure shows two communities identified. The color gradient denotes the centrality of the communities (betweenness). Node size is relative to the centrality (betweenness) of each feature.
(C) UMAP clustering of cells and their associated cell types generated from scRNA-seq data published by Zhang et al. (2020). Expression of top essential genes identified in (A), in various cell types, is shown in subsequent UMAPs.