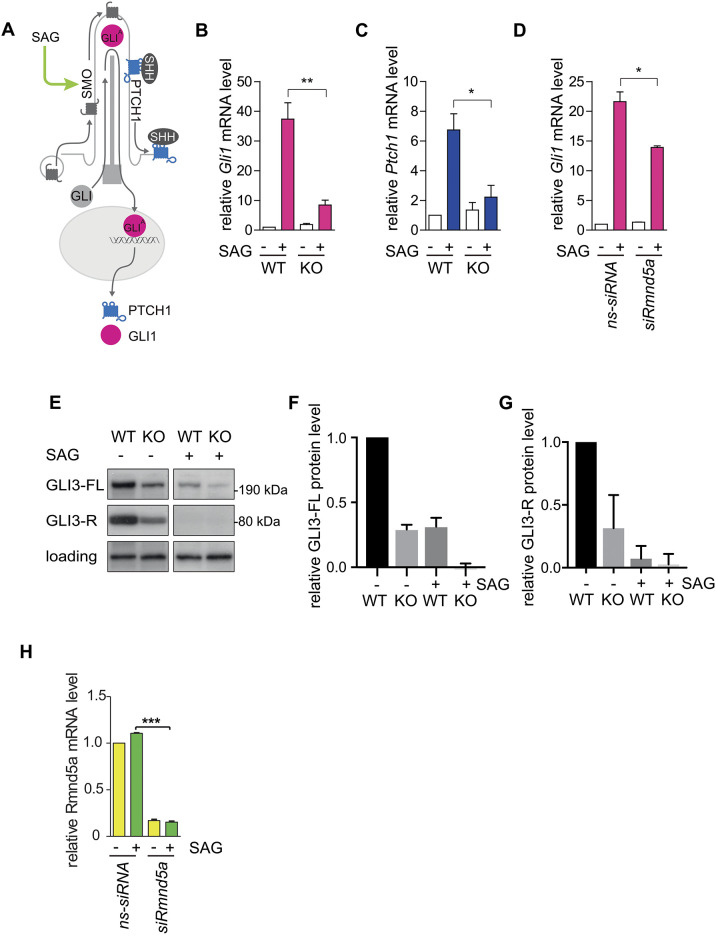
Fig. 3.
GID deficiency interferes with SHH signaling. (A) Schematic representation of primary cilium-dependent SHH signaling in the ‘on state’. After binding of SHH ligand to PTCH1, it relieves the inhibition of smoothened (SMO). Then, GLI transcription factors are activated (GLIA), turning on expression of downstream genes, such as Gli1 and Ptch1. (B–D) qPCR of two SHH signaling markers (Gli1 in B,D; Ptch1 in C) during RMND5A KO (B,C) or knockdown (D). Cells were cultured under cilia-induced conditions (high glucose DMEM, 0.5% FCS) with or without SAG (100 nM) treatment for 24 h and harvested for further analysis. ns-siRNA, control siRNA; siRmnd5a, RMND5A knockdown. Mean±s.e.m., n=3. *P<0.05; **P<0.01 (two-tailed, unpaired Student's t-test). (E) Western blot of GLI3 (same blot and exposure time). WT and KO cells were cultured under cilia-induced conditions (high glucose DMEM, 0.5% FCS) with or without SAG (100 nM) treatment for 24 h and harvested. A protein that cross-reacted with the GLI3 antibody was used as a loading control. (F,G) Quantification of western blots as in E showing relative protein level of GLI3 full-length form (F) and of GLI3 repressor form (G) in NIH-3T3 WT and RMND5A KO cells. Signal intensities of all lanes were measured on the same western blot with same exposure times and are normalized to the WT −SAG condition. WT −SAG, GLI3-FL/GLI3-R=1.51; KO −SAG, GLI3-FL/GLI3-R=1.4. Data are mean±s.e.m. of n=3. (H) qPCR of Rmnd5a mRNA during RMND5A knockdown. Cells were cultured under cilia-induced conditions (high glucose DMEM, 0.5% FCS) with or without SAG (100 nM) treatment for 24 h and harvested. Data are mean±s.e.m. of n=3. ***P<0.001 (two-tailed, unpaired Student's t-test). Knockdown efficiency showed an 85% reduction of Rmnd5a mRNA.