Abstract
Objective
This study evaluates the probiotic activity of three vaginal Lactobacillus gasseri (H59.2, IMAUFB014, and JCM1131) and one non-vaginal L. plantarum ATCC14917 against three Candida albicans (ATCC10231, candidiasis, and healthy vaginal microbiota). Displacement of lactobacilli and adhesion inhibition of C. albicans were evaluated on an abiotic surface through adhesion assays with different experimental settings (ES) through low (1.0E + 03 CFU/ml) and high (1.00E + 09 CFU/ml) levels of colonization. ES simulated dysbiosis (ES1 and ES4), candidiasis (ES2), and healthy vaginal microbiota (ES3).
Results
At ES2 and ES3, L. gasseri H59.2 showed discrepant inhibition values among C. albicans isolates (ES2: P = 0.008, ES3: P = 0.030; two‐way ANOVA). L. plantarum was only displaced by 23%, 31%, 54%, and 94% against low and high levels of C. albicans ATCC10231. L. plantarum was less displaced, when compared to L. gasseri strains (ES1: 61–84%, ES2: 82–96%, ES3: 83–95%, and ES4: 73–97%), showing multiple statistical differences (ES1: P = < 0.001, ES2: P = 0.003, and ES3: P = < 0.001; two‐way ANOVA). L. plantarum also showed a superior inhibition of C. albicans ATCC10231 in ES1 (81%) and ES2 (58%) when compared to L. gasseri strains (ES1: 27–73%, P < 0.001; and ES2:1–49%, P < 0.001; two‐way ANOVA).
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-022-06114-z.
Keywords: Candida albicans, Initial adhesion, Lactobacillus gasseri, Lactobacillus plantarum, Probiotics
Introduction
The vaginal microbiota is colonized by several microorganisms [1], where commensal Lactobacillus species act as defense mechanism [2]. Lactobacilli can adhere and biosynthesize antimicrobial compounds reducing colonization by pathogens [1] associated with bacterial vaginosis, aerobic vaginitis, and candidiasis [3]. Thus, probiotics could be an efficient alternative to antimicrobial treatment, which reduces commensal microbiota and increases resistance [4, 5]. Lactobacilli may restore healthy microbiota, as postulated by Mitrea and colleagues [5]. Although Candida spp. is commensal, this genus can become an opportunistic pathogen in high levels [6, 7] leading to vulvovaginal candidiasis and evolving in more serious urinary tract infections and venereal diseases [8, 9]. Studies reported the application of lactobacilli biofilms and biosurfactants against pathogens [10–13]. However, these approaches are designed to treat established infections and do not endure in vaginal microbiota [14]. Another approach could be the colonization of the mucosal epithelia by new and more probiotic lactobacilli [15], allowing a permanent integration in the microbiota. However, the inhibition of the initial adhesion of pathogens by lactobacilli is not fully understood [16, 17]. It is important to compare the variability of vaginal and non-vaginal lactobacilli to inhibit the adhesion of pathogens and to avoid their displacement. This study evaluated the probiotic ability of three vaginal L. gasseri and one non-vaginal L. plantarum to protect an abiotic surface in initial adhesion assays, assessing the lactobacilli displacement and the inhibition of three C. albicans through different scenarios of dysbiosis conditions, candidiasis, and healthy vaginal microbiota.
Main text
Methods
From previous studies [3, 18], three Lactobacillus gasseri (H59.2, IMAUFB014, and JCM1131), two Candida albicans (one isolate from a healthy vaginal microbiota, and another from candidiasis), and C. albicans ATCC10231 and L. plantarum ATCC14917 were used in this study. Lactobacillus strains were grown in Man, Rogosa and Sharpe agar for 48 h at 37 °C under microaerophilic conditions [19, 20]. C. albicans strains were grown in Sabouraud Dextrose Agar at 37 °C for 18 h [19–21]. Brain Heart Infusion (BHI) broth was used for initial adhesion assays [20].
Initial adhesion assays
Each microorganism was concentrated in 5 ml of sterile phosphate-buffered saline (PBS) solution. Both suspensions were collected by centrifugation (4000 g, 12 min, at room temperature), and washed twice with PBS. The pellet was resuspended according to the growth curves to 1.0E + 03 colony-forming unit (CFU)/ml and 1.00E + 09 CFU/ml (Additional files 1 and 2) by optical density at 600 nm (OD600). Four experimental settings (ES) were made from concentration combinations (Additional file 3), varying on low levels of lactobacilli (1.00E + 03 CFU/ml) against low and high levels of C. albicans (ES1 and ES2, respectively) and then on high levels of lactobacilli (1.00E + 09 CFU/ml) against low and high levels of C. albicans (ES3 and ES4, respectively). These ES mimicked dysbiosis conditions (ES1 and ES4), candidiasis (ES2), and healthy vaginal microbiota (ES3). Initial adhesion assays were realized using a preincubation of lactobacilli for 4 h at 37 °C with 120 rpm [22, 23] and then evaluating the initial adhesion of Candida albicans with the pre-adhered lactobacilli during 30 min at same conditions [23–26], as illustrated in the flowchart (Additional file 4) [27, 28]. Non-adherent microorganisms were removed by PBS washing. All experimental assays were repeated three times on different days.
Microscopy analysis and cell quantification
After adhesion assay, a PBS washing step was carried out on coverslips, which were fixed with ethanol (96%; v/v) and stained with crystal violet at 3% for 1 min [29]. From each coverslip, 15 random fields were photographed in Olympus BX50 microscope under 1000x [23, 30] using the AmScope MU633-FL camera. The number of Lactobacillus spp. and C. albicans were counted from each picture (Additional file 5), being expressed as the number of cells per glass surface ± standard deviation (Additional file 6 and Additional file 7).
Statistical analysis
The statistical analysis was realized through two-tailed ANOVA (ANalysis Of VAriance) with post-hoc Tukey HSD (Honestly Significant Difference) and Student t-test using JASP software version 0.13. ANOVA analysis evaluated differences in and between ES, post-hoc Tukey HSD test analyzed differences between species on the same ES, and Student t-test assessed differences between samples and controls. P values ≤ 0.050 were statistically significant.
Results
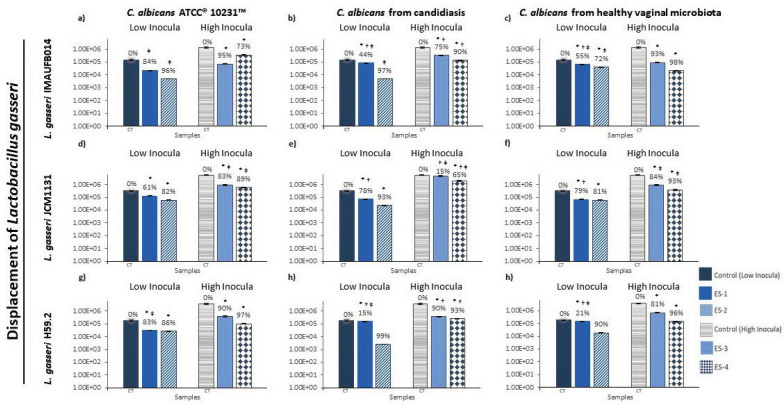
Initial adhesion assays were realized through different experimental settings, simulating dysbiosis conditions (ES1 and ES4), candidiasis (ES2), and healthy vaginal microbiota (ES3). The lactobacilli displacement was evaluated on low levels (ES1 and ES2) against low and high concentrations of C. albicans and then on high levels (ES3 and ES4), as shown in Additional file 6. On low levels of lactobacilli, the displacement was between 15 and 99% (see Fig. 1). At ES1, C. albicans ATCC10231 induced bigger displacement of L. gasseri IMAUFB014 (84%; P = 0.010, two‐way ANOVA) and H59.2 (83%; P < 0.001, two‐way ANOVA) showing significant differences among C. albicans (Tukey's post hoc, P < 0.05). Likewise, all C. albicans showed to be statistically different in their displacement ability among the L. gasseri. C. albicans from healthy vaginal microbiota was able to displace 99% of L. gasseri IMAUFB014, while C. albicans isolated from candidiasis demonstrated 99% of displacement against L. gasseri JCM1131. At ES2, no significant differences were found in the displacement among L. gasseri. At ES3, C. albicans isolated from candidiasis showed statistical differences, evidencing a greater ability to displace L. gasseri H59.2 (90%; P < 0.001, two‐way ANOVA). L. gasseri JCM1131 showed only 15% of displacement by C. albicans isolated from candidiasis, being statistically different when compared to C. albicans ATCC10231 (83%; P = 0.001, Tukey's post hoc) and C. albicans isolated from healthy vaginal microbiota (84%; P < 0.001, Tukey's post hoc). At ES4, L. gasseri JCM1131 showed 65% of displacement by C. albicans isolated from candidiasis, but it only evidenced a significant difference against C. albicans isolated from healthy vaginal microbiota (93%; P = 0.045, Tukey's post hoc).
Fig. 1.
Displacement of Lactobacillus gasseri by Candida albicans obtained through initial adhesion assays. Displacement of L. gasseri by C. albicans after initial adhesion treatments with the experimental setting of high and low inoculum in the glass surface. The percentage of adhesion of L. gasseri is the result of the variation in the adhesion of L. gasseri and C. albicans strains to coverslip in comparison to controls (CT, 100% of adhesion) when incubated alone at the same conditions. Statistical analysis: *P < 0.05 when using t-student statistical analysis (95% confidence interval) for comparison of lactobacilli control and sample tested in the adhesion assay; †P < 0.05 analyzed using two-tailed ANOVA statistical test (95% confidence interval) for comparison of displacement values from all lactobacilli strains induced by a certain C. albicans isolate tested in the adhesion assay; ‡P < 0.05 analyzed using two-tailed ANOVA statistical test (95% confidence interval) for comparison of displacement values from a certain strain of lactobacilli among all C. albicans isolates tested in the adhesion assay
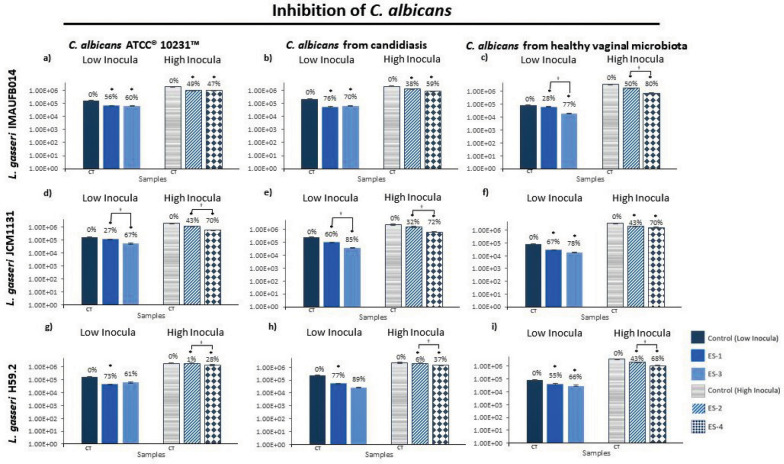
The adhesion inhibition of C. albicans by L. gasseri was also evaluated (see Fig. 2). At ES1, L. gasseri JCM1131 and L. gasseri IMAUFB014 showed statistical differences among C. albicans (L. gasseri JCM1131 P = 0.006, and L. gasseri IMAUFB014 P = 0.002; two‐way ANOVA). L. gasseri JCM1131 evidenced the lowest inhibition rate against C. albicans ATCC10231 (27%), illustrating significant values when compared against C. albicans isolated from candidiasis (60%; P = 0.016, Tukey's post hoc) and C. albicans isolated from healthy vaginal microbiota (67%; P = 0.006, Tukey's post hoc). While L. gasseri IMAUFB014 showed a more efficient inhibition rate against C. albicans isolated from candidiasis (76%; P = 0.002, Tukey's post hoc). At ES2, L. gasseri H59.2 was the only strain to show statistically inhibition values among C. albicans isolates (P = 0.008; two‐way ANOVA). Again, at ES3, only L. gasseri H59.2 demonstrated a statistical difference in its inhibition ability (P = 0.030; two‐way ANOVA) against C. albicans ATCC10231 (61%) and C. albicans isolated from candidiasis (89%; P = 0.034, Tukey's post hoc). At ES4, all C. albicans showed significant inhibition rates (C. albicans ATCC10231: P = 0.010; C. albicans isolated from candidiasis: P = 0.011; C. albicans isolated from healthy microbiota: P = 0.025, two‐way ANOVA analysis). L. gasseri IMAUFB014 showed the highest inhibition rate against C. albicans isolated from healthy microbiota (80%), being statistically different to C. albicans ATCC10231 (47%; P = 0.016, Tukey's post hoc).
Fig. 2.
The probiotic activity of Lactobacillus gasseri against Candida albicans showed in initial adhesion assays. Inhibition of C. albicans by L. gasseri after initial adhesion treatments with the experimental setting of high and low inoculum in the glass surface. The percentage of adhesion of C. albicans is the result of the variation in the adhesion of L. gasseri and C. albicans strains to coverslip in comparison to controls (CT, 100% of adhesion) when incubated alone at the same conditions. Statistical analysis: *P < 0.05 when using t-student statistical analysis (95% confidence interval) for comparison of candida control and sample tested in the adhesion assay; †P < 0.05 analyzed using two-tailed ANOVA statistical test (95% confidence interval) for comparison of inhibition values between experimental setting (ES) for each evaluated C. albicans isolated in the adhesion assay
Probiotic ability of Lactobacillus plantarum ATCC14917 was realized against C. albicans ATCC10231 and compared with L. gasseri evidencing significant displacement and inhibition values (see Additional file 7 and Fig. 3A). The displacement values of L. plantarum were 23% and 54% against low (ES1) and high (ES2) levels of C. albicans, respectively. These values were significantly inferior to L. gasseri (ES1: 61–99% and ES2: 82–96%), more exactly: L. gasseri IMAUFB014 (ES1: P < 0.001and ES2: P = 0.002, Tukey's post hoc); L. gasseri JCM1131 (ES1: P = 0.003 and ES2: P = 0.025, Tukey's post hoc); and L. gasseri H59.2 (ES1: P < 0.001 and ES2: P = 0.012, Tukey's post hoc). At ES3, L. plantarum was only displaced by 31% evidencing again a better resistance when compared to L. gasseri (ES3: 83–95%), specifically: L. gasseri IMAUFB014 (P < 0.001, Tukey's post hoc); L. gasseri H59.2 (P < 0.001, Tukey's post hoc); and L. gasseri JCM1131 (P = 0.001, Tukey's post hoc). At ES4, L. plantarum was displaced 94% without statistical differences. As shown in Fig. 3B, the adhesion inhibition of C. albicans by L. plantarum demonstrated a superior activity in ES1 (81%) and ES2 (58%) when compared to L. gasseri (ES1:27–73% and ES2:1–49%; both P < 0.001, two‐way ANOVA). At ES3 and ES4, L. plantarum showed the lowest inhibition rate (50–56%) without statistical differences.
Fig. 3.
Preliminary analysis of the displacement of Lactobacillus plantarum by Candida albicans and its probiotic activity on C. albicans through initial adhesion assays. A Displacement of L. plantarum ATCC 14917 by C. albicans ATCC 10231 after initial adhesion treatments with the experimental setting of high and low inoculum in the glass surface. Statistical analysis: *P < 0.05 when using t-student statistical analysis (95% confidence interval) for comparison of lactobacilli control and sample tested in the adhesion assay; †P < 0.05 analyzed using two-tailed ANOVA statistical test (95% confidence interval) for comparison of displacement values between L. plantarum and L. gasseri strains in the adhesion assay at same experimental setting. B Inhibition of C. albicans by L. plantarum after initial adhesion treatments with the experimental setting of high and low inoculum in the glass surface. Statistical analysis: *P < 0.05 when using t-student statistical analysis (95% confidence interval) for comparison of candida control and sample tested in the adhesion assay; †P < 0.05 analyzed using two-tailed ANOVA statistical test (95% confidence interval) for comparison of inhibition values between experimental setting (ES) for each evaluated C. albicans ATCC 10231 isolated in the adhesion assay. No statistically significant values were found
Discussion
The use of Lactobacillus is a low-risk alternative for antimicrobial resistance and its adverse effects [31]. It is well-known that a prerequisite for C. albicans' pathogenicity is the initial adhesion to host cells [32] before leading to genital or urinary tract infections [8, 9]. This study focused on the intrinsic probiotic activity of lactobacilli against C. albicans. Although studies reported lactobacilli biofilm/biosurfactant activities against pathogens [10–13], few authors evaluated the inhibition of the initial adhesion of pathogens [16, 17, 23, 33]. Some studies previously characterized the inhibition of the initial adhesion of certain pathogens, such as Gardnerella vaginalis, Prevotella bivia, Mobiluncus mulieris [23], Listeria monocytogenes [34], and Streptococcus mutans [17]. Recently, He et al. [35] evaluated the probiotic activities of Lactobacillus species on the inhibition of the initial adhesion of several pathogens. However, only L. gasseri demonstrated a more probiotic activity against C. albicans, when compared to L. crispatus. Our results agreed with He et al. [35], reporting differences in probiotic activity among Lactobacillus species/strains against C. albicans isolates. However, there is still scarce information about how these Candida albicans can be inhibited by Lactobacillus strains/species or even to displace different lactobacilli.
An alternative approach could be used by the colonization of new and more probiotic lactobacilli in this environment [15], being permanently assimilate in the vaginal microbiota. Once incorporated, certain lactobacilli species should be able to produce supernatant and eventually evolve in biofilm formation [15, 36], such as L. plantarum. So, the initial adhesion is a vital step for the human epithelial colonization and the inhibition of pathogens being worthy to study.
The present study evaluated three L. gasseri as inherent vaginal lactobacilli and a single L. plantarum (ATCC 14917) as a strong probiotic species atypical of the vaginal microbiota. Our results evidenced statistical differences between the displacement values of L. gasseri strains by the same C. albicans isolate and the variability of each Lactobacillus to inhibit different C. albicans. This variability agrees with a study realized by De Gregorio et al. [11] with Lactobacillus crispatus on the adhesion of Candida species, which evidenced the strain-specific probiotic activity of L. crispatus. So, it is plausible to assume that the remaining Lactobacillus species could also evidence discrepancies in their probiotic ability and displacement resistance against different C. albicans isolates, as proposed by Zangl et al. [15]. The application of different lactobacilli from other biological sources in the human epithelial colonization could increment the probiotic activity of the remaining commensal microbiota, as suggested in other studies [37–39]. These studies together with our results of low displacement values in L. plantarum against low or normal levels of C. albicans suggested the potential application of non-human lactobacilli to sustain a more resilient healthy microbiota. L. plantarum ATCC 14917 demonstrated high inhibition percentages of C. albicans ATCC 10231, being more efficient and statistically different when compared to L. gasseri. Our results surpassed the rates of inhibition on C. albicans reported by Dos Santos et al. [12]. Further studies should evaluate longitudinal colonization between non-vaginal lactobacilli and vaginal lactobacilli against Candida species in vitro assays.
Limitations
There are some major limitations in this study: (1) it is a preliminary study realized on an abiotic surface and unable to establish an efficient report on human epithelial colonization; (2) the study did not evaluate the longitudinal colonization between lactobacilli and C. albicans; and (3) the study only compared the probiotic activity of L. plantarum against a single Candida albicans.
Supplementary Information
Additional file 1: Figure S1. Comparison of growth calibration curves between Lactobacillus gasseri strains and Lactobacillus plantarum ATCC 14917 used in this study.
Additional file 2: Figure S2. Comparison of growth calibration curves between C. albicans strains used in this study.
Additional file 3: Figure S3. Representation of the experimental settings in adhesion assays simulating dysbiosis conditions (ES1 and ES4), candidiasis (ES2), and healthy vaginal microbiota (ES3).
Additional file 4: Figure S4. Illustration of the flowchart on the procedures used in the initial adhesion assays of the present study. The flowchart was realized using the online software CmapTools (https://cmap.ihmc.us/) [28].
Additional file 5: Figure S5. Comparison of sample for L. gasseri IMAUFB014 against C. albicans ATCC 10231 observed in the OLYMPUS BX50 microscope for each experimental setting (ES), evaluating the initial adhesion of C. albicans and the displacement of pre-adhered lactobacilli during 30 min. A Random field (1000x) of L. gasseri IMAUFB014 (1.00E + 03 CFU/ml) against C. albicans ATCC 10231 (1.00E + 03 CFU/ml) at ES1. B Random field (1000x) of L. gasseri IMAUFB014 (1.00E + 03 CFU/ml) against C. albicans ATCC 10231 (1.00E + 09 CFU/ml) at ES2. C Random field (1000x) of L. gasseri IMAUFB014 (1.00E + 09 CFU/ml) against C. albicans ATCC 10231 (1.00E + 03 CFU/ml) at ES3. D Random field (1000x) of L. gasseri IMAUFB014 (1.00E + 09 CFU/ml) against C. albicans ATCC 10231 (1.00E + 09 CFU/ml) at ES4.
Additional file 6: Table S1. Displacement of Lactobacillus gasseri by Candida albicans obtained through initial adhesion assays.
Additional file 7: Table S2. Displacement of Lactobacillus plantarum by Candida albicans obtained through initial adhesion assays.
Acknowledgements
A special recognition deserves all colleagues of the Microbiology Institute of USFQ and COCIBA, as well as the Research Office of Universidad San Francisco de Quito and COCIBA for their financial support in this study.
Abbreviations
- MRS
De Man, Rogosa and Sharpe agar
- SDA
Sabouraud dextrose agar
- BHI
Brain heart infusion
- PBS
Phosphate buffered saline
- CFU
Colony-forming unit
- ES
Experimental setting
- ANOVA
Analysis of variance
- Tukey HSD
Tukey honestly significant difference test
Author contributions
AM was responsible for the study and analysis design. Conceptualization, methodology, formal analysis, data curation, writing was done by RJRA; BOGA; DEM-V and AM. The reviewing manuscript was realized by RJRA and AM. Research project supervision was conducted by AM in the Microbiology Institute USFQ. All the authors read and approved the final manuscript.
Funding
This work was supported by COCIBA Research Grant 2018–2019 through project ID: 12260 entitled “Adhesión inicial y resistencia antimicrobiana de Candida sp. aisladas de la microbiota humana”, under regulations of the Ministry of Health of Ecuador (Contrato Marco de Acceso a los Recursos Genéticos No. MAE-DNB-CM-2016-0046).
Availability of data and materials
All the data supporting the study findings are within the manuscript. Additional detailed information and raw data are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participants
No human participants, human material, or human data were involved in this study.
Consent for publication
Not applicable.
Competing interests
The authors have no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Robert Josue Rodríguez-Arias and Bryan Omar Guachi-Álvarez contributed equally to this work.
Contributor Information
Robert Josue Rodríguez-Arias, Email: rrodrigueza@alumni.usfq.edu.ec.
Bryan Omar Guachi-Álvarez, Email: bguachi@estud.usfq.edu.ec.
Dominique Esther Montalvo-Vivero, Email: dominique.montalvo@alumni.usfq.edu.ec.
António Machado, Email: amachado@usfq.edu.ec, Email: machadobq@gmail.com.
References
- 1.Borges S, Silva J, Teixeira P. The role of lactobacilli and probiotics in maintaining vaginal health. Arch Gynecol Obstet. 2013;289:479–489. doi: 10.1007/s00404-013-3064-9. [DOI] [PubMed] [Google Scholar]
- 2.Hickey RJ, Zhou X, Pierson JD, Ravel J, Forney LJ. Understanding vaginal microbiome complexity from an ecological perspective. Transl Res. 2012;160:267–282. doi: 10.1016/j.trsl.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salinas AM, Osorio VG, Herrera DP, Vivanco JS, Trueba AF, Machado A. Vaginal microbiota evaluation and prevalence of key pathogens in Ecuadorian women: an epidemiologic analysis. Sci Rep. 2020 doi: 10.1038/s41598-020-74655-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lannitti T, Palmieri B. Therapeutical use of probiotic formulations in clinical practice. Clin Nutr. 2010;29:701–725. doi: 10.1016/j.clnu.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitrea L, Călinoiu LF, Precup G, Bindea M, Rusu B, Trif M, et al. Inhibitory potential of Lactobacillus plantarum on Escherichia coli. Bull Univ Agric Sci Vet Med Cluj Napoca Food Sci Technol. 2017;74:99. [Google Scholar]
- 6.Allonsius CN, Vandenheuvel D, Oerlemans EFM, Petrova MI, Donders GGG, Cos P, et al. Inhibition of Candida albicans morphogenesis by chitinase from Lactobacillus rhamnosus GG. Sci Rep. 2019 doi: 10.1038/s41598-019-39625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitrea L, Ranga F, Fetea F, Dulf FV, Rusu A, Trif M, et al. Biodiesel-derived glycerol obtained from renewable biomass—a suitable substrate for the growth of Candida zeylanoides yeast strain ATCC 20367. Microorganisms. 2019;7:265. doi: 10.3390/microorganisms7080265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behzadi P, Behzadi E, Ranjbar R. Urinary tract infections and Candida albicans. Cent Eur J Urol. 2015;68:96–101. doi: 10.5173/ceju.2015.01.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behzadi P, Behzadi E, Pawlak-Adamska EA. Urinary tract infections (UTIs) or genital tract infections (GTIs)? It’s the diagnostics that count. GMS Hyg Infect Control. 2022;2019(14):1–12. doi: 10.3205/dgkh000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jalilsood T, Baradaran A, Song AAL, Foo HL, Mustafa S, Saad WZ, et al. Inhibition of pathogenic and spoilage bacteria by a novel biofilm-forming Lactobacillus isolate: a potential host for the expression of heterologous proteins. Microb Cell Fact. 2015;14:1–14. doi: 10.1186/s12934-015-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Gregorio PR, Parolin C, Abruzzo A, Luppi B, Protti M, Mercolini L, et al. Biosurfactant from vaginal Lactobacillus crispatus BC1 as a promising agent to interfere with Candida adhesion. Microb Cell Fact. 2020;19:1–16. doi: 10.1186/s12934-020-01390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dos Santos CI, França YR, Campos CDL, Bomfim MRQ, Melo BO, Holanda RA, et al. Antifungal and antivirulence activity of vaginal Lactobacillus spp. Products against Candida vaginal isolates. Pathogens. 2019;8:1–20. doi: 10.3390/pathogens8030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez S, Garcia JG, Williams R, Elmassry M, West A, Hamood A, et al. Lactobacilli spp.: Real-time evaluation of biofilm growth. BMC Microbiol. 2020;20:1–9. doi: 10.1186/s12866-020-01753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Cerbo A, Palmieri B, Aponte M, Morales-Medina JC, Iannitti T. Mechanisms and therapeutic effectiveness of lactobacilli. J Clin Pathol. 2016;69:187–203. doi: 10.1136/jclinpath-2015-202976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zangl I, Pap IJ, Aspöck C, Schüller C. The role of lactobacillus species in the control of candida via biotrophic interactions. Microb Cell. 2020;7:1–14. doi: 10.15698/mic2020.01.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machado A, Almeida C, Salgueiro D, Henriques A, Vaneechoutte M, Haesebrouck F, et al. Fluorescence in situ hybridization method using peptide nucleic acid probes for rapid detection of Lactobacillus and Gardnerella spp. BMC Microbiol. 2013;13:82. doi: 10.1186/1471-2180-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda Y, Cho O, Sugita T, Ogishima D, Takeda S. Culture supernatants of Lactobacillus gasseri and L. crispatus inhibit Candida albicans biofilm formation and adhesion to HeLa cells. Mycopathologia. 2018;183:691–700. doi: 10.1007/s11046-018-0259-4. [DOI] [PubMed] [Google Scholar]
- 18.Pacha-Herrera D, Vasco G, Cruz-betancourt C, Galarza JM, Barragán V, Machado A. Vaginal microbiota evaluation and Lactobacilli quantification by qPCR in pregnant and non-pregnant women : a pilot study. Front Cell Infect Microbiol. 2020;10:1–13. doi: 10.3389/fcimb.2020.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribeiro FC, de Barros PP, Rossoni RD, Junqueira JC, Jorge AOC. Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J Appl Microbiol. 2017;122:201–211. doi: 10.1111/jam.13324. [DOI] [PubMed] [Google Scholar]
- 20.Matsubara VH, Wang Y, Bandara HMHN, Mayer MPA, Samaranayake LP. Probiotic lactobacilli inhibit early stages of Candida albicans biofilm development by reducing their growth, cell adhesion, and filamentation. Appl Microbiol Biotechnol. 2016;100:6415–6426. doi: 10.1007/s00253-016-7527-3. [DOI] [PubMed] [Google Scholar]
- 21.Vilela SFG, Barbosa JO, Rossoni RD, Santos JD, Prata MCA, Anbinder AL, et al. Lactobacillus acidophilus ATCC 4356 inhibits biofilm formation by C. albicans and attenuates the experimental candidiasis in Galleria mellonella. Virulence. 2015;6:29–39. doi: 10.4161/21505594.2014.981486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishiyama K, Seto Y, Yoshioka K, Kakuda T, Takai S, Yamamoto Y, et al. Lactobacillus gasseri SBT2055 reduces infection by and colonization of Campylobacter jejuni. PLoS ONE. 2014;9:e108827. doi: 10.1371/journal.pone.0108827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machado A, Jefferson KK, Cerca N. Interactions between Lactobacillus crispatus and bacterial vaginosis (BV)-associated bacterial species in initial attachment and biofilm formation. Int J Mol Sci. 2013;14:12004–12012. doi: 10.3390/ijms140612004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castro J, Henriques A, Machado A, Henriques M, Jefferson KK, Cerca N. Reciprocal interference between Lactobacillus spp. and Gardnerella vaginalis on initial adherence to epithelial cells. Int J Med Sci. 2013;10:1193–8. doi: 10.7150/ijms.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fidel PL, Barousse M, Espinosa T, Ficarra M, Sturtevant J, Martin DH, et al. An intravaginal live Candida challenge in humans leads to new hypotheses for the immunopathogenesis of vulvovaginal candidiasis. Infect Immun. 2004;72:2939–2946. doi: 10.1128/IAI.72.5.2939-2946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seneviratne CJ, Samaranayake LP, Ohshima T, Maeda N, Jin LJ. Identification of antifungal molecules from novel probiotic Lactobacillus bacteria for control of Candida infection. Hong Kong Med J. 2016;22(7):34–6. [PubMed] [Google Scholar]
- 27.Behzadi P, Gajdács M. Writing a strong scientific paper in medicine and the biomedical sciences: a checklist and recommendations for early career researchers. Biol Future. 2021;72:395–407. doi: 10.1007/s42977-021-00095-z. [DOI] [PubMed] [Google Scholar]
- 28.Novak JD, Cañas AJ. The Theory Underlying Concept Maps and How to Construct and Use Them, Technical Report IHMC CmapTools 2006–01 Rev 01–2008. IHMC C. 2008:1–36. http://www.ode.state.or.us/teachlearn/subjects/science/resources/msef2010-theory_underlying_concept_maps.pdf%5Cnpapers://dee23da0-e34b-4588-b624-f878b46d7b3d/Paper/p348. Accessed 24 May 2022.
- 29.Weerasekera MM, Wijesinghe GK, Jayarathna TA, Gunasekara CP, Fernando N, Kottegoda N, et al. Culture media profoundly affect Candida albicans and Candida tropicalis growth, adhesion and biofilm development. Mem Inst Oswaldo Cruz. 2016;111:697–702. doi: 10.1590/0074-02760160294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chauviere G, Coconnier M-H, Kerneis S, Fourniat J, Servin AL. Adhesion of human Lactobacillus acidophilus strain LB to human enterocyte-like Caco-2 cells. J Gen Microbiol. 1992;138:1689–1696. doi: 10.1099/00221287-138-8-1689. [DOI] [PubMed] [Google Scholar]
- 31.Jeavons HS. Prevention and treatment of vulvovaginal candidiasis using exogenous Lactobacillus. J Obstet Gynecol Neonatal Nurs. 2003;32:287–296. doi: 10.1177/0884217503253439. [DOI] [PubMed] [Google Scholar]
- 32.Graf K, Last A, Gratz R, Allert S, Linde S, Westermann M, et al. Keeping Candida commensal: How lactobacilli antagonize pathogenicity of Candida albicans in an in vitro gut model. Dis Model Mech. 2019;12:1–16. doi: 10.1242/dmm.039719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alves P, Castro J, Sousa C, Cereija TB, Cerca N. Gardnerella vaginalis outcompetes 29 other bacterial species isolated from patients with bacterial vaginosis, using in an in vitro biofilm formation model. J Infect Dis. 2014;210:593–596. doi: 10.1093/infdis/jiu131. [DOI] [PubMed] [Google Scholar]
- 34.Lezzoum-Atek S, Bouayad L, Hamdi TM. Influence of some parameters on the ability of Listeria monocytogenes, Listeria innocua, and Escherichia coli to form biofilms. Vet World. 2019;12:459–465. doi: 10.14202/vetworld.2019.459-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Y, Niu X, Wang B, Na R, Xiao B, Yang H. Evaluation of the inhibitory effects of Lactobacillus gasseri and Lactobacillus crispatus on the adhesion of seven common lower genital tract infection-causing pathogens to vaginal epithelial cells. Front Med. 2020 doi: 10.3389/fmed.2020.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alves R, Barata-Antunes C, Casal M, Brown AJP, van Dijck P, Paiva S. Adapting to survive: how Candida overcomes host-imposed constraints during human colonization. PLoS Pathog. 2020;16:e1008478. doi: 10.1371/journal.ppat.1008478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hasslöf P, Hedberg M, Twetman S, Stecksén-Blicks C. Growth inhibition of oral mutans streptococci and candida by commercial probiotic lactobacilli - an in vitro study. BMC Oral Health. 2010 doi: 10.1186/1472-6831-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wasfi R, Abd El-Rahman OA, Zafer MM, Ashour HM. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J Cell Mol Med. 2018;22:1972–83. doi: 10.1111/jcmm.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, He J, Pan D, Wu Z, Guo Y, Zeng X, et al. Metabolomics analysis of Lactobacillus plantarum ATCC 14917 adhesion activity under initial acid and alkali stress. PLoS ONE. 2018;13:e0196231. doi: 10.1371/journal.pone.0196231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Comparison of growth calibration curves between Lactobacillus gasseri strains and Lactobacillus plantarum ATCC 14917 used in this study.
Additional file 2: Figure S2. Comparison of growth calibration curves between C. albicans strains used in this study.
Additional file 3: Figure S3. Representation of the experimental settings in adhesion assays simulating dysbiosis conditions (ES1 and ES4), candidiasis (ES2), and healthy vaginal microbiota (ES3).
Additional file 4: Figure S4. Illustration of the flowchart on the procedures used in the initial adhesion assays of the present study. The flowchart was realized using the online software CmapTools (https://cmap.ihmc.us/) [28].
Additional file 5: Figure S5. Comparison of sample for L. gasseri IMAUFB014 against C. albicans ATCC 10231 observed in the OLYMPUS BX50 microscope for each experimental setting (ES), evaluating the initial adhesion of C. albicans and the displacement of pre-adhered lactobacilli during 30 min. A Random field (1000x) of L. gasseri IMAUFB014 (1.00E + 03 CFU/ml) against C. albicans ATCC 10231 (1.00E + 03 CFU/ml) at ES1. B Random field (1000x) of L. gasseri IMAUFB014 (1.00E + 03 CFU/ml) against C. albicans ATCC 10231 (1.00E + 09 CFU/ml) at ES2. C Random field (1000x) of L. gasseri IMAUFB014 (1.00E + 09 CFU/ml) against C. albicans ATCC 10231 (1.00E + 03 CFU/ml) at ES3. D Random field (1000x) of L. gasseri IMAUFB014 (1.00E + 09 CFU/ml) against C. albicans ATCC 10231 (1.00E + 09 CFU/ml) at ES4.
Additional file 6: Table S1. Displacement of Lactobacillus gasseri by Candida albicans obtained through initial adhesion assays.
Additional file 7: Table S2. Displacement of Lactobacillus plantarum by Candida albicans obtained through initial adhesion assays.
Data Availability Statement
All the data supporting the study findings are within the manuscript. Additional detailed information and raw data are available from the corresponding author on reasonable request.