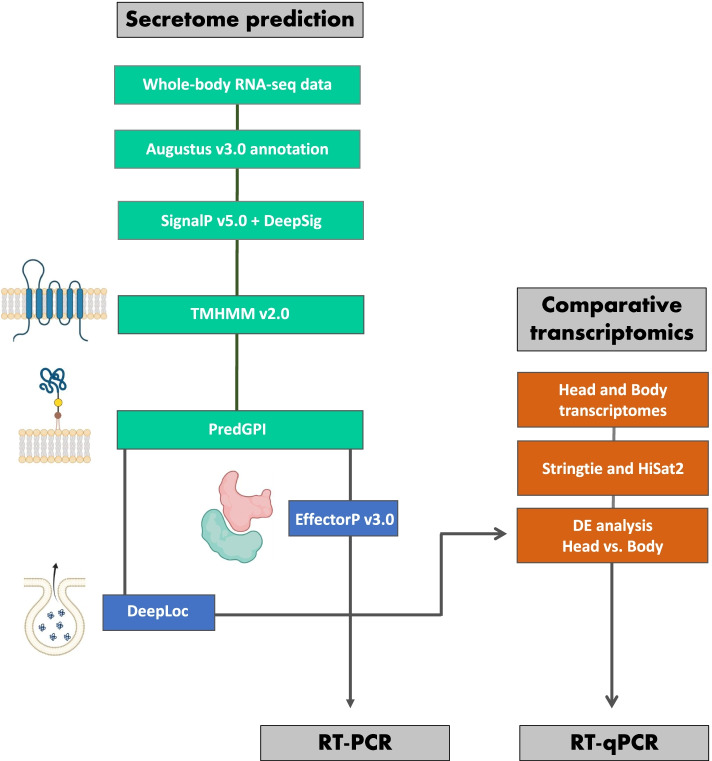
Fig. 1.
Diagrammatic representation of the methodology used to predict putative effectors from Diuraphis. noxia whole-body transcriptome data. RNA-seq data was mined for protein sequences with characteristics of extracellularly secreted proteins. This data was also analysed with EffectorP and combined into a single list, constituting the secretome. Selected sequences were analysed for salivary gland specific expression through RT-PCR. Orthologs of the putatively secreted D. noxia sequences were identified in three aphid species. The differential expression of the orthologs between the head versus body transcriptome data of three aphid species was determined to identify transcripts with a Log2-FC>2. This predicted differential expression was confirmed in D. noxia head, body and salivary gland tissue through RT-qPCR analysis