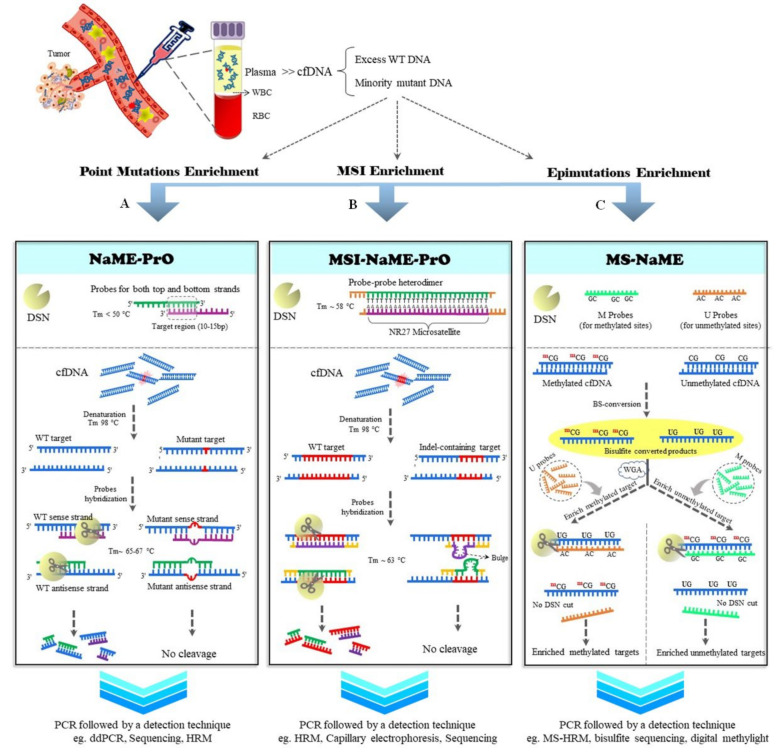
Figure 1.
Applications of nuclease-assisted minor-allele enrichment (NaME) method in cell-free DNA (cfDNA) or genomic DNA: (A) Workflow for NaME using probe overlap (NaME-PrO) for enrichment of mutations. The overlapping probes are designed to match sense and antisense strands of wild-type (WT) targets. The nucleotides within the probe overlap region determine the target sequence. Following denaturation step and probe hybridization, duplex-specific nuclease (DSN) preferentially digests fully matched double-stranded sequences, thereby retaining intact the mutated strands; (B) workflow for enrichment of indel-containing sequences to enhance detection of microsatellite instability (MSI-NaME-PrO). The overlapping probes for targeting microsatellite NR27, as an example, are illustrated. The polyA homopolymer has a low Tm, such that the probe–probe and the probe–target hybrids show a ~5 °C difference. Following probe hybridization, DSN digests WT alleles, while indel-containing sequences remain substantially undigested because of bulges within probe–target hybrids; (C) workflow of methylation-sensitive NaME (MS-NaME): U probes are designed to bind uracil-containing sequences (which are generated via bisulfite-mediated cytosine deamination). M probes are designed to bind 5mC-containing DNA. Following denaturation and whole-genome amplification (WGA), either U probes or M probes are applied to enrich methylated and unmethylated targets, respectively.