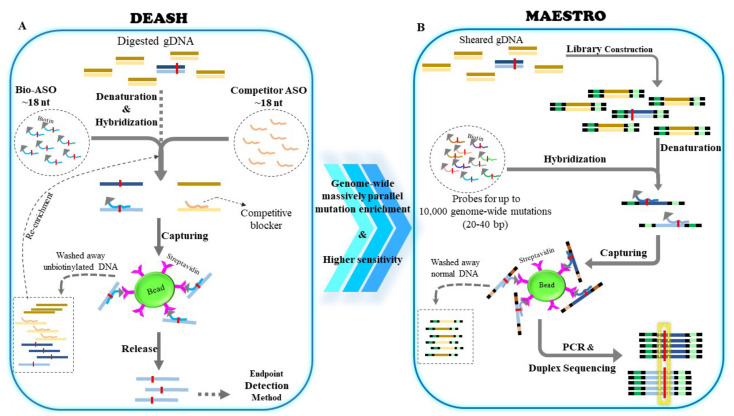
Figure 3.
(A) Principles of DNA enrichment via allele-specific hybridization (DEASH), and minor-allele-enriched sequencing through recognition oligonucleotides (MAESTRO). The biotinylated allele-specific oligonucleotide (bio-ASO) is designed to bind mutant strands, while competitor ASO attaches to wild-type strands. Following denaturation and hybridization, the bio-ASO/target hybrids are captured by streptavidin-coated beads. The unbound DNA can undergo further cycles of DEASH. The enriched single-stranded targets are eluted and purified; (B) workflow for minor-allele-enriched sequencing through recognition oligonucleotides (MAESTRO) technique. The underlying principle is similar to DEASH, although MAESTRO operates without competitive blockers and provides genome-wide massively parallel enrichment. Following NGS library construction and barcoding top and bottom DNA strands, the biotinylated probes complementary for up to 10,000 genome-wide mutations are employed to bind mutant strands. Afterwards, the mutation-containing strands are preferentially captured by streptavidin-coated beads, are amplified, and undergo duplex sequencing.