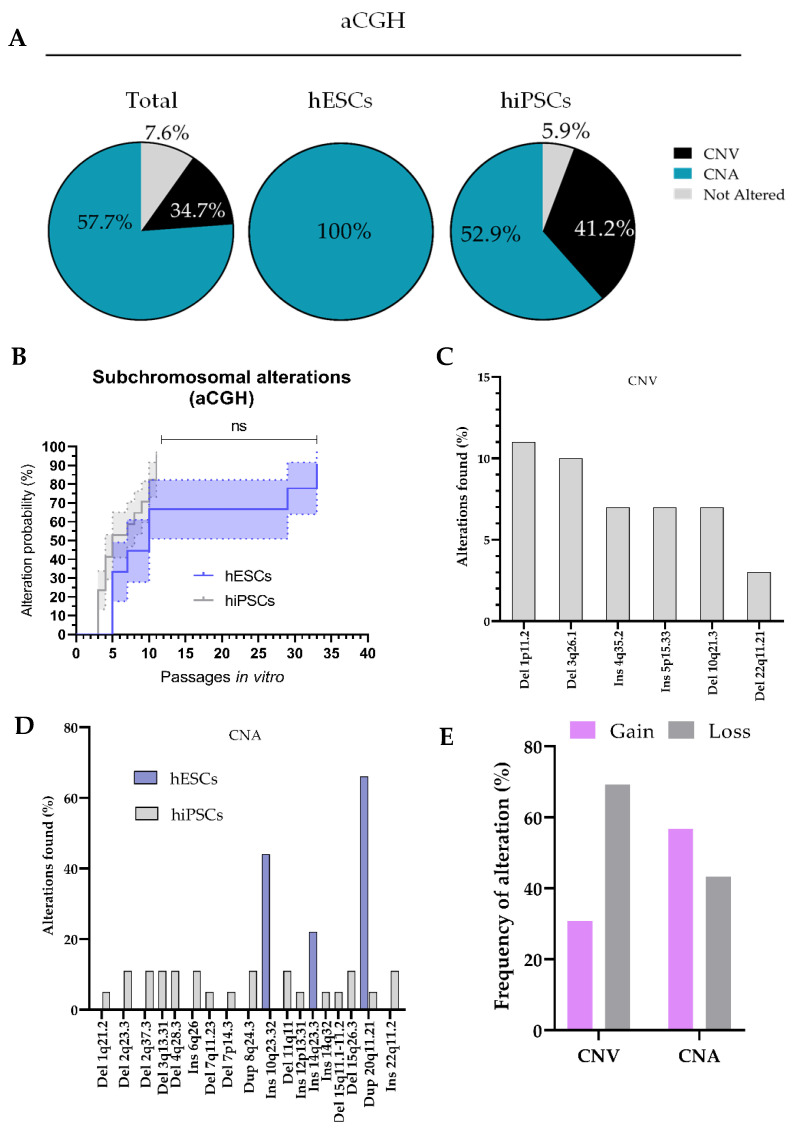
Figure 5.
Recurrent subchromosomal Copy Number Variations (CNVs) and pathogenic Copy Number Alterations (CNAs) are detected in cultured hESC s and hiPSCs by Comparative Genomic Hybridization CGH array (aCGH) analysis: (A) Pie charts showing the percentage of cells harboring Copy Number Variations (CNVs) or Copy Number Alterations (CNAs), as well as cell lines for which no alteration were detected; (B) survival analysis (Kaplan–Meier curves) of the evolution of the probability (%) of hESCs and hiPSCs being affected by subchromosomal genomic alterations detected by aCGH over time (passages in vitro). Error bars represent standard error (S.E.M.); (C,D) histograms showing the frequency at which different types of CNVs (C) and CNAs (D) are found in analyzed hESCs and hiPSCs; (E) bar graph showing the frequency at which DNA gains and losses are detected with aCGH for both CNV and CNA group.