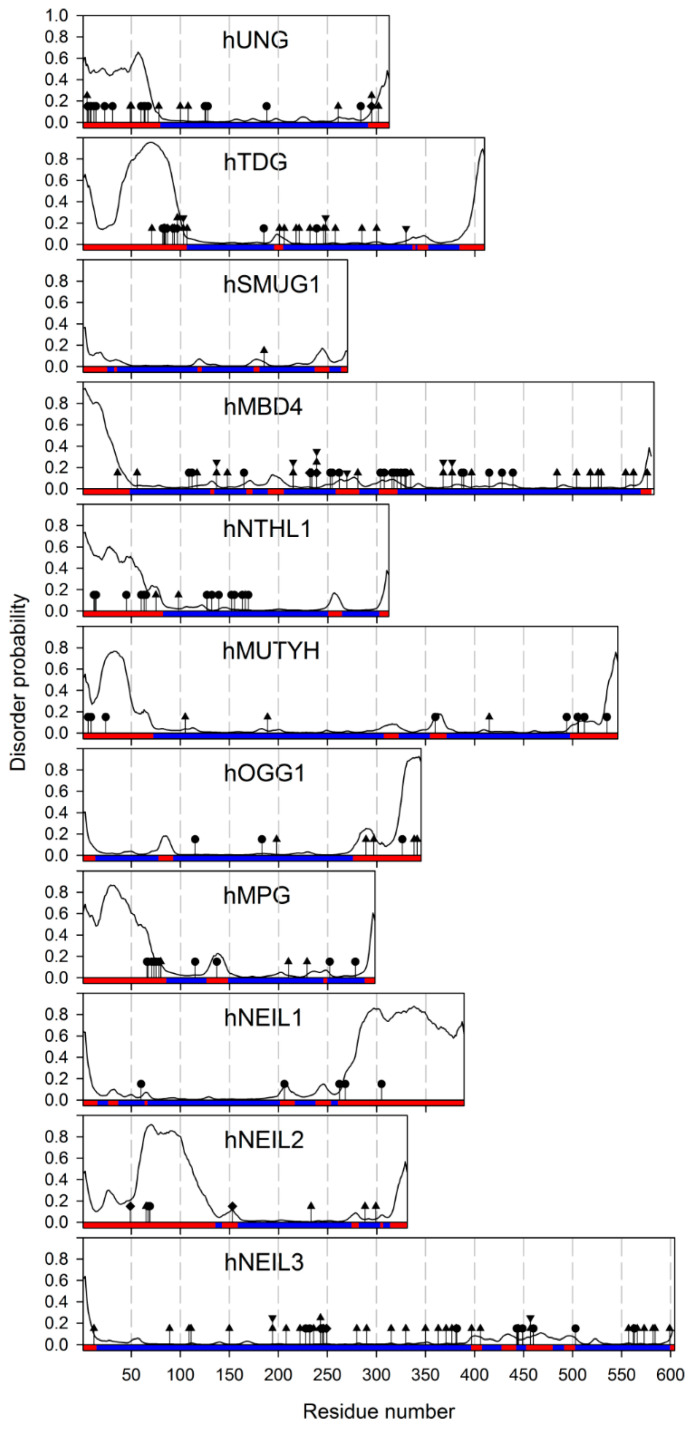
Figure 2.
Predicted disorder and sites of post-translational modifications in human DNA glycosylases. The disorder probabilities were calculated using the ESpritz neural network [12]. The colored bar corresponds to ESpritz predictions: ordered (blue) and disordered (red). The sites of post-translational modifications are labeled by circles (Ser/Thr/Tyr phosphorylation), diamonds (Lys acetylation), triangles (Lys ubiquitylation), and reverse triangles (Lys sumoylation). The sites of modifications are taken from the PhosphoSitePlus proteomic database [13] and low-throughput studies discussed in the main text.