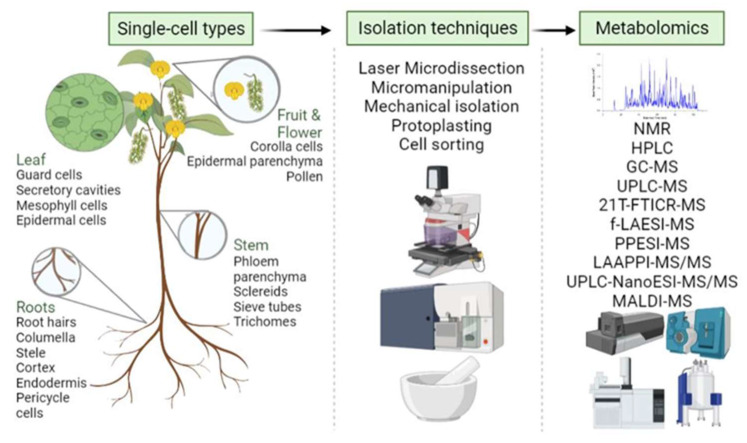
Figure 5.
Single-cell metabolomics workflow from different cell types (e.g., epidermal cells, guard cells, root hairs, floral organ cells, root tip stem cells, root nodule cells, hairy root culture cells, in vitro callus cultures, parenchyma and sclerenchyma cells, mesophyll and bundle sheath cells, and inner endodermal cells), and organelles including apoplast, chloroplast, and mitochondria. They were isolated from plant tissues using methods including laser microdissection, laser-ablation, laser capture microdissection-liquid vortex capture, micromanipulation, mechanical isolation, protoplasting, pressure probe, and cell sorting. NMR: Nuclear magnetic resonance; HPLC: High performance liquid chromatography; LMD LC-MS: Laser Microdissection liquid-chromatography mass spectrometry; LAAPPI: Laser ablation atmospheric pressure photoionization; GC: Gas chromatography; LAESI: Laser ablation electrospray ionization; picoPPESI: picolitre pressure-probe electrospray-ionization; fLAESI: Optical fiber-based laser ablation electrospray ionization; 21TFTICR-MS: 21 tesla (T) Fourier transform ion cyclotron resonance MS; LSC-MS: Live single-cell MS; UPLC: Ultra performance liquid chromatography; NanoESI: Nano electrospray ionization; MALDI: Matrix-assisted laser desorption and ionization.