Figure 2. The HBB and G6PD polymorphisms located on chromosome chr11 and chr X, respectively.
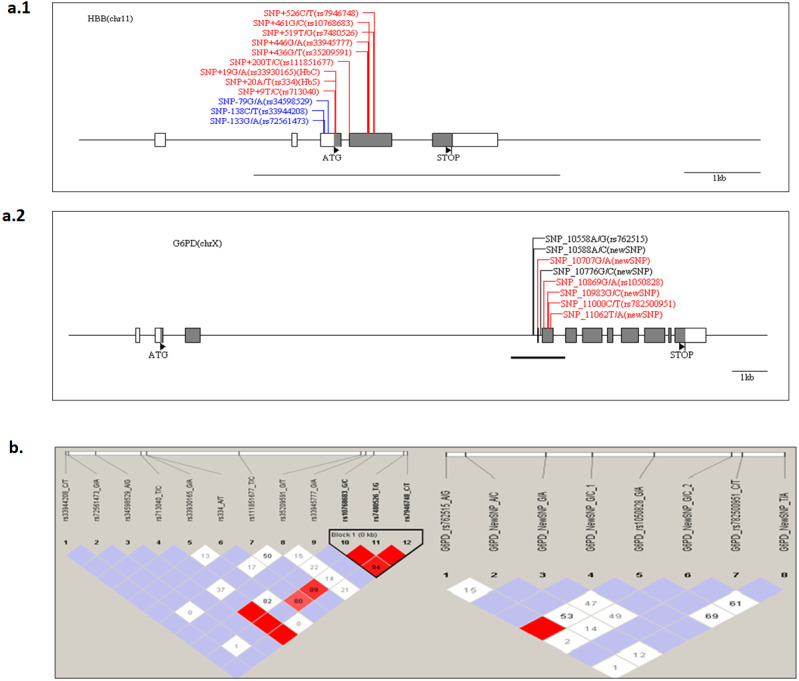
The Haploview software provides the linkage disequilibrium map of HBB and G6PD genes. (A) The coding and UTR regions are indicated by black and white rectangles, respectively, for each loci. SNP positions are numbered per the initiation codon ATG considered as +1 (indicated by a black triangle). The genomic sequence used for alignment is GenBank sequence; reference genes were NG_009015.2 and NG_059281 for HBB and G6PD, respectively. The different colors are: label blue: SNP ‘promoter’ region, label ‘Black’, SNP‘intronic’ region and label ‘Red’, ‘exonic’ region. An underlined black line shows the boundaries of PCR amplicon in each gene. (B) The linkage disequilibrium map of LD plots within regions of 800 and 700 bases for HBB and G6PD genes, respectively. The LD plots show pairwise D′ values given in diamonds for each statistical comparison between the SNPs. The different shades of color represent D′ values (between 0 and 1). In white and red ‘diamond’, the values of D′ are indicated, an empty red diamond indicates that D′ > 0.97.
