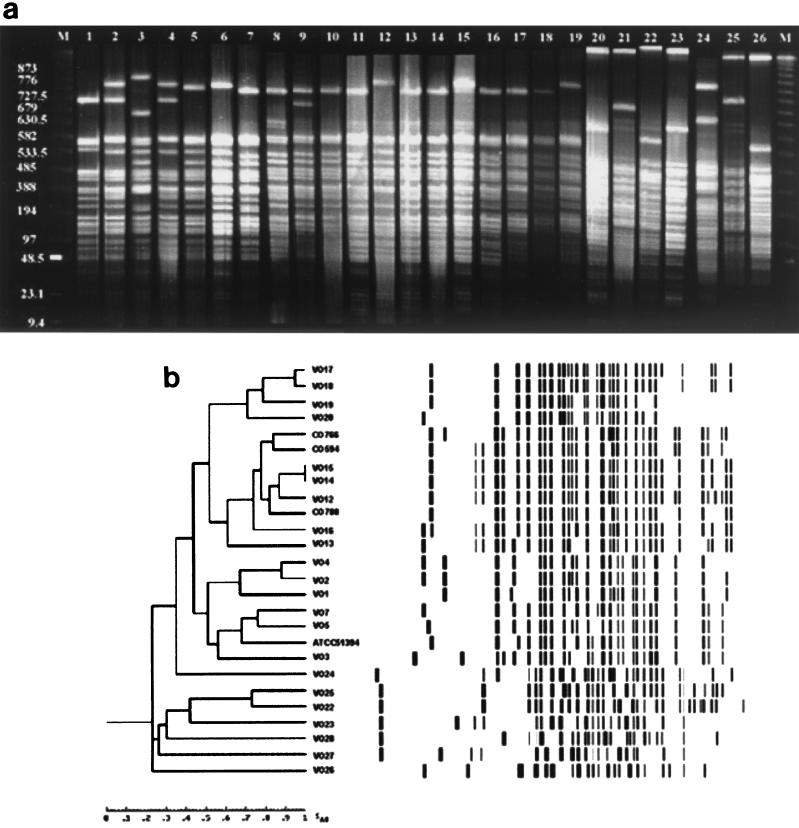
FIG. 4.
(a) Fingerprint patterns obtained from PFGE of SfiI-digested DNA of clinical and environmental V. cholerae O1, O139, and non-O1, non-O139 isolates. Lanes M, 1-kb molecular weight ladder; lanes 1 to 3 and 12, O1 strains VO1, VO2, VO3, and VO13, respectively; lanes 4 to 6, O1 strains VO4, VO5, and VO7, respectively; lanes 7 to 11 and 13 to 15, O139 strains MO45 (= ATCC 51394), CO594, CO766, CO788, VO12, VO14, VO15, and VO16, respectively; lanes 16 to 19, O139 strains VO17, VO18, VO19, and VO20, respectively; lanes 20, and 21, non-O1, non-O139 strains VO22, and VO23, respectively; lanes 22 to 26, non-O1, non-O139 strains VO24, VO25, VO26, VO27, and VO28, respectively; (b) Digitized PFGE analysis of SfiI-digested profiles obtained from genomic DNA of clinical and environmental V. cholerae O1, O139, and non-O1, non-O139 isolates. The Dendrogram was generated by using the average percentages of matched bands summarizing the degrees of similarity of the SfiI restriction patterns of genomic DNA of V. cholerae O1, O139, and non-O1, non-O139 strains.