Abstract
Parkinson’s disease (PD) is generally considered a sporadic disorder, but a strong genetic background is often found. The aim of this study was to identify the underlying genetic cause of PD in two affected siblings and to subsequently assess the role of mutations in Cathepsin B (CTSB) in susceptibility to PD. A typical PD family was identified and whole-exome sequencing was performed in two affected siblings. Variants of interest were validated using Sanger sequencing. CTSB p.Gly284Val was genotyped in 2077 PD patients and 615 unrelated healthy controls from the Czech Republic, Ireland, Poland, Ukraine, and the USA. The gene burden analysis was conducted for the CTSB gene in an additional 769 PD probands from Mayo Clinic Florida familial PD cohort. CTSB expression and activity in patient-derived fibroblasts and controls were evaluated by qRT-PCR, western blot, immunocytochemistry, and enzymatic assay. The CTSB p.Gly284Val candidate variant was only identified in affected family members. Functional analysis of CTSB patient-derived fibroblasts under basal conditions did not reveal overt changes in endogenous expression, subcellular localization, or enzymatic activity in the heterozygous carrier of the CTSB variant. The identification of the CTSB p.Gly284Val may support the hypothesis that the CTSB locus harbors variants with differing penetrance that can determine the disease risk.
Keywords: Parkinson’s disease, familial forms, monogenic forms, CTSB, fibroblasts
1. Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder [1]. Clinical symptoms include bradykinesia, resting tremors, muscular rigidity, and a good response to levodopa or dopamine-agonist treatment [2]. The degeneration of dopaminergic neurons in the substantia nigra and the accumulation of α-synuclein are the main pathological hallmarks of PD [3]. It is mostly a sporadic disease, but about 10% of cases are familial [4]. Mutations in PD-related genes are usually found in patients with early-onset PD (autosomal recessive) or in patients with positive family history (autosomal dominant) [4]. In addition to the well-described causative genes, many more have been identified as potential risk factors. The most recent genome-wide association study (GWAS) revealed almost 100 susceptibility loci [5]. Although the genetics of PD in the Polish population have been explored, known genetic causes are not commonly observed [6]. This suggests that there may be specific PD-associated genetic variants within unresolved loci in the Polish population. Understanding the genetic causes of PD within families can be informative for the more frequent sporadic form of the disease. There are a number of examples in the genetics of PD that show variable penetrance at a single locus, e.g., SNCA and LRRK2, which were both identified through family-based studies but later found to harbor common variants of intermediate/low penetrance. Herein, we describe a genetic investigation of a Polish kindred with two siblings affected by PD and basic functional evaluation of the nominated CTSB mutant in patient-derived fibroblasts.
2. Results
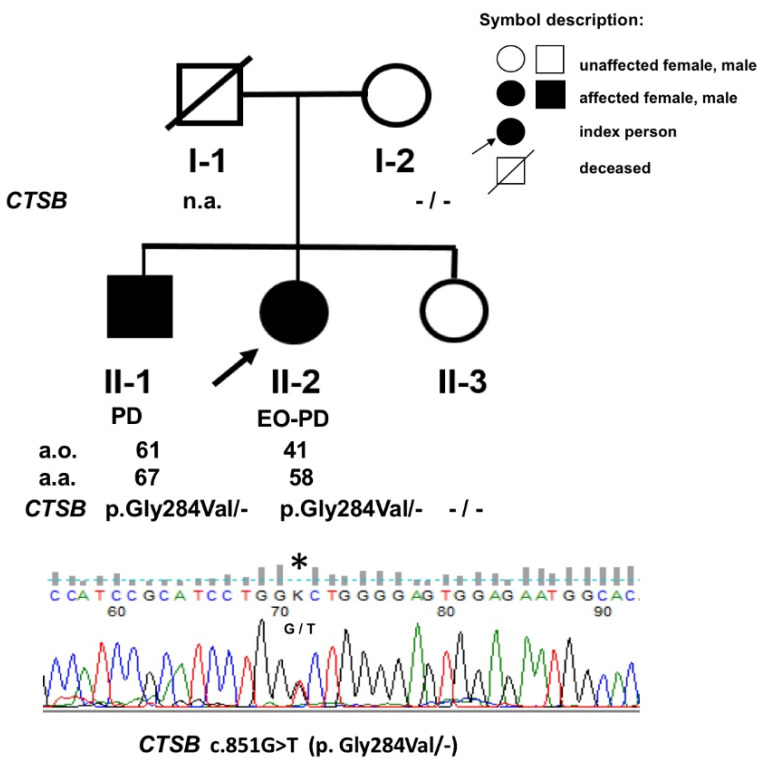
The proband was a 58-year-old female (II-2) with 17 years history of PD (Figure 1). She developed right-hand rigidity and was responsive to levodopa treatment. After 11 years of disease onset, she developed on/off fluctuations and bilateral deep brain stimulation to the subthalamic nucleus was implanted. Her brother (Figure 1) (II-1) developed a right-hand tremor at the age of 61 and was then diagnosed with PD. After 6 years of disease onset, he continued to show a good levodopa response (Supplementary Video S1) [7]. No other known history of PD was found within the family, due to limited information from siblings who currently live in Germany.
Figure 1.
Pedigree of the Polish PD family chosen for the study. (n.a.—not available, a.o.—age of disease onset, a.a.—age of patient’s analysis, *—the mutation site).
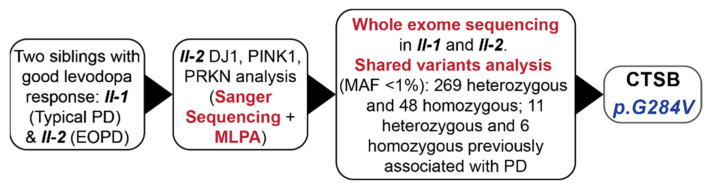
The analysis of the three main EOPD genes—PRKN, PINK1, and DJ1 (exons sequencing and MLPA) did not show the presence of pathogenic variants (Figure 2). The WES analysis performed for both affected subjects (II-1, II-2) identified 317 rare (MAF < 1%) missense or loss-of-function variants shared between the sibling pair. Of the 317 variants, 269 were heterozygous and 48 variants were homozygous (Supplementary Table S1). Seventeen of the identified variants, including 11 heterozygous and 6 homozygous, were previously reported to be associated with PD (Table 1).
Figure 2.
Flowchart of genetic experiments conducted on the typical PD family. EOPD = early onset Parkinson’s disease, MAF = minor allele frequency, MLPA—Multiplex ligation-dependent probe amplification.
Table 1.
Summary of shared variants in affected siblings previously associated with PD.
| Position (GRCh38) | rs | Ref | Alt | CADD Score | Classification | Gene | Evidence |
|---|---|---|---|---|---|---|---|
| Homozygous | |||||||
| chr6:g.32518589 | rs147439581 | C | T | 13.6 | Nonsyn SNV | HLA-DRB5 | GWAS [5] |
| chr6:g.32519397 | rs112872773 | C | G,T | 10.9 | Nonsyn SNV | HLA-DRB5 | GWAS [5] |
| chr6:g.32521967 | rs780328684 | G | TGG,- | Frameshift Del, Frameshift Sub | HLA-DRB5 | GWAS [5] | |
| chr6:g.32584262 | rs150747106 | C | T,G | 21.9 | Nonsyn SNV | HLA-DRB1 | GWAS [8] |
| chr10:g.17849736 | rs71497225 | G | C | 8.1 | Nonsyn SNV | MRC1 | Decrease the risk of PD [9] |
| chr10:g.17849710 | rs71497223 | A | G | 6.7 | Nonsyn SNV | MRC1 | Decrease the risk of PD [9] |
| Heterozygous | |||||||
| chr1:g.13782630 | rs140700877 | A | C | 22.7 | Nonsyn SNV | PRDM2 | SNP more frequent observed in PD [10] |
| chr2:g.182756345 | rs138065612 | C | T | 24.1 | Nonsyn SNV | DNAJC10 | Decrease the risk of PD [11] |
| chr3:g.52378778 | rs201064587 | G | A | 21.2 | Nonsyn SNV | DNAH1 | SNP more frequent observed in PD [12] |
| chr8:g.11845732 | Novel | C | A | 26.7 | Nonsyn SNV | CTSB | Confirmed PD/LBD GWAS loci [5,13] |
| chr9:g.87637944 | rs200255856 | G | A | 32 | Nonsyn SNV | DAPK1 | Positive impact on LRRK2 and synuclein expression in animal models [14] |
| chr10:g.23104162 | rs2296466 | A | G | 15.56 | Nonsyn SNV | MSRB2 | Important in regulation of mitophagy [15] |
| chr11:g.113387894 | rs35657708 | G | T | 10.01 | Nonsyn SNV | ANKK1 | SNP more frequent observed in PD [16] |
| chr12:g.122340779 | Novel | T | A | 17.03 | Nonsyn SNV | CLIP1 | Increase risk of PD in LRRK2 patients [17] |
| chr15:g.78593232 | rs76071148 | T | A | 15.39 | Nonsyn SNV | CHRNA5 | Decrease risk of PD [18] |
| chr16:g.1443261 | Novel | C | T | 36 | Nonsyn SNV | CCDC154 | Higher level of protein observed in serum of PD patients [19] |
| chr19:g.55185948 | rs140748270 | G | C | 23 | Nonsyn SNV | PTPRH | Positive association in single WES study (1000 patients) [20] |
Alt—alternative, CADD—Combined Annotation Dependent Depletion, Chr—chromosome, GWAS—genome wide association studies.
The variant CTSB p.Gly284Val (c.851G > T) was identified as the top variant responsible for the symptoms and disease within the family. Interestingly, the CTSB locus was previously identified as a GWAS target, and this variant is predicted as likely pathogenic according to in silico analysis ((MutationTaster 2021, Berlin Institute of Health, Berlin, Germany), SIFT4G (Bioinformatics Institute, Singapore), CADD v1.4 (University of Washington, Hudson-Alpha Institute for Biotechnology, St. Louis, MO, USA) score 26.7) [5]. To further validate our findings, 5 independent cohorts (Table 2) were screened with no additional carriers for CTSB p.Gly284Val found. The result from gene burden analysis was negative when using the Mayo Clinic Biobank cohort and the Mayo Clinic familial PD cohort (Supplementary Table S2).
Table 2.
Basic demographic characteristic of the population genotyped for CTSB c.851G > T (p.Gly284Val) variant.
| Country of Origin | PD (N Females) | Age of Onset (Mean ± SD) | Controls (N Females) |
|---|---|---|---|
| USA | 997 (358) | 65.2 (±11.9) | - |
| Poland | 610 (250) | 59.3 (±12.6) | 248 (125) |
| Ireland | 320 (142) | 57.0 (±11.9) | 343 (217) |
| Ukraine | 122 (57) | 59.9 (±11.6) | 24 (12) |
| Czech Republic | 28 (14) | 59.3 (±11.8) | - |
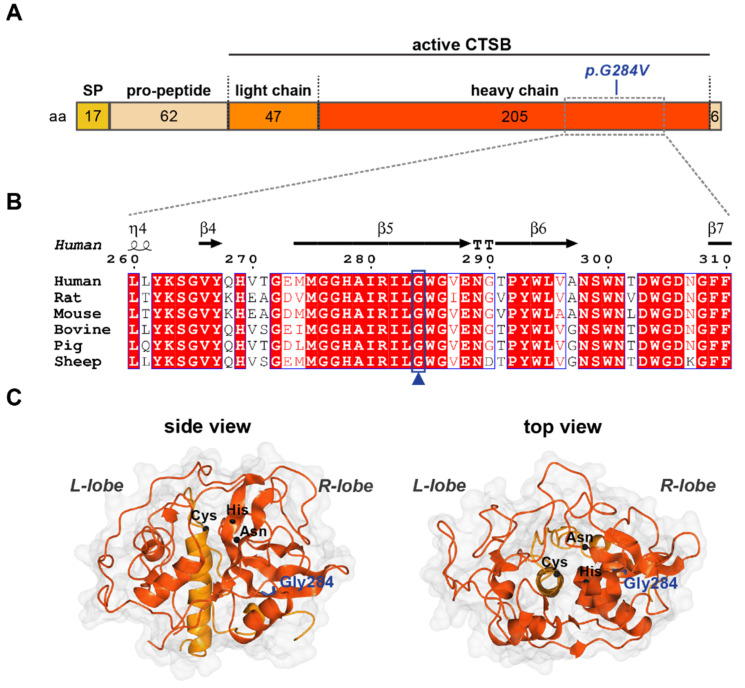
To address the potential significance of the p.Gly284Val substitution, we first inspected the surrounding sequence and the localization of the residue within the 3D structure of the protein (Figure 3). CTSB is synthesized as a preproenzyme which, upon cleavage of the N-terminal signal and pro-peptide regions, is further processed into the mature, active protease consisting of light and heavy chains (Figure 3A). Although Gly284 does not directly localize to the active site of the enzyme, it resides within a highly conserved region of the heavy chain that folds into the R-lobe of CTSB and may modulate catalytic activity (Figure 3B,C).
Figure 3.
Glycine 284 is conserved in CTSB across species. (A) Primary structure of CTSB indicating length of fragments in amino acids (aa) and position of the p.Gly284V mutation. SP—signal peptide. (B) Structure-informed multiple sequence alignment of six CTSB homologs. The secondary structure for human CTSB (PDB: 6AY2) is shown above. Boxed residues are conserved: white background with red text indicates functionally equivalent residues; red background with white text indicates sequence conservation. The blue box and arrowhead highlight Gly284, which is conserved across all species analyzed. (C) Crystal structure of mature human CTSB (PDB: 6AY2) shown in surface/ribbon representation, indicating the left (L) and right (R) lobes which constitute the mature enzyme. Heavy and light chains are colored red-orange and orange, respectively. Gly284 (blue) is shown in cylindrical representation; C-alphas of the catalytic triad: Cys (29) His (199) and Asn (219) are depicted as black spheres.
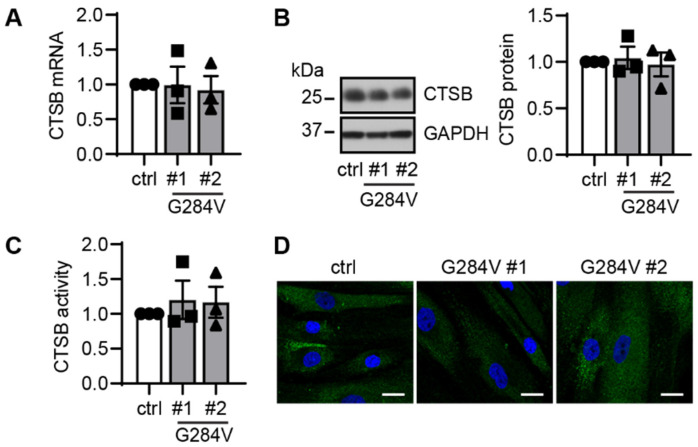
To functionally characterize the novel mutation, we examined mRNA and protein expressions as well as its activity and the localization using patient-derived fibroblasts from both PD patients and age-matched controls (Figure 4). During culture, there were no apparent differences in cell proliferation, morphology, or survival observed between patients and controls. Under standard, non-stress conditions, heterozygous mutant fibroblasts showed no major differences in total CTSB transcript or protein levels (Figure 4A,B). There was also no significant change in enzymatic activity of CTSB (Figure 4C). Immunocytochemistry showed similar lysosomal distribution of the CTSB signal between patients and control fibroblasts (Figure 4D).
Figure 4.
Functional analysis of CTSB in patient-derived skin cells. (A) qRT-PCR showed comparable mRNA levels in both CTSB mutant fibroblasts and the controls cells. (B) Representative western blot images of control and two CTSB mutant fibroblast lines (left). Western blot quantification showed similar levels of total CTSB protein levels in both CTSB mutant, and the WT control cells (right). (C) CTSB activity assay using cell lysates showed no differences in total enzymatic activity in the mutant fibroblast compared to the control. Circle—control, square—patient #1 carrying CTSB G284V, triangle—patient #2 carrying CTSB G284V. (D) Representative images of CTSB immunofluorescence staining (green) in fibroblasts at baseline condition. Scale bar: 20 µm. n = 3 independent experiments. Data are normalized to set control as 1 and are shown as mean with standard error. One-way ANOVA.
3. Discussion
Our study identified a novel, rare CTSB p.Gly284Val variant in the affected family that may play a role in the pathogenesis of PD. Although we did not show differences in CTSB gene burden analysis in small cohorts of control and PD cases, accumulating evidence from recent large genetic studies has revealed a strong involvement of CTSB in PD development. The CTSB locus was identified as a genetic risk locus in a recent GWAS analysis [5]. CTSB was considered as PD expression quantitative trait loci in the transcriptomic analysis [21]. The CTSB variant rs1293298 was also identified as the modifier of risk and age of onset in GBA associated PD and Lewy body dementia [22]. On the other hand, we conducted CTSB gene burden analysis and CTSB p.Gly284Val genotyping which did not find any associations. In a limited number of functional studies of CTSB in PD, the direct involvement of CTSB in α-synuclein degradation was revealed. CTSB and CTSL were shown to jointly cleave α-synuclein within its amyloid region and circumvent fibril formation [13]. However, CTSB also contributed to the generation of C-terminally truncated α-synuclein species in symptomatic SNCA p.Ala53Thr transgenic mice [23]. Yet, the impact of the CTSB variant in PD patients on protein activity had never been investigated before. A structure-informed multiple sequence alignment of human CTSB against a selection of homologs found that Gly284 is evolutionarily conserved and lies within the heavy chain of active enzymes [24]. This indicates the potential structural importance of this residue, thus a mutation such as p.Gly284Val may adversely affect protein function.
Cathepsins are lysosomal proteases that are mainly found in the acidic compartments where they are most active, but each cathepsin has its own specific optimal pH environment [25]. Cathepsins take part in different physiological and pathological processes and play critical roles in intracellular protein degradation, energy metabolism, and immune response [26]. The importance of the lysosomal pathway in PD pathogenesis was described previously, and cathepsins belong to the most crucial lysosomal proteins [27]. Significant reduction in the lysosomal degradation capacity, substantially enlarged lysosomes, and increased lysosome number were observed in the CTSB and CTSL double knockout human neuroblastoma cells [28]. Moreover, CTSB knockout cells exhibited accumulated lysosomal protein LAMP1 (lysosomal-associated membrane protein 1) [28]. Lack of CTSB was also shown to impair lysosomal trafficking during neural development [29]. Additionally, CTSB was found to indirectly control the transcription factor EB (TFEB), which is the most important regulator of autophagy and lysosomal gene expression [30].
For functional evaluation of the variant, we did not find clear evidence supporting haploinsufficiency in the fibroblasts at basal condition. However, CTSB activity is also context-dependent, and the impact of the variant may be different in neurons or other models. Thus, a more thorough analysis of the CTSB variant in relevant cells of the brain and/or under disease-relevant stress conditions may be needed to uncover potential functional deficits. Lysosomal dysfunction has a well-established role in the pathogenesis of PD. Currently, there are several ongoing clinical trials targeting this pathway (for example NCT02914366, NCT04127578). Therefore, further analysis of lysosomal dysfunction in PD may result in the development of potential new therapeutic targets.
4. Materials and Methods
4.1. Clinical Examination
Four members of mixed origins, a Polish-German family (Figure 1) with typical PD from southern Poland, were recruited from the Department of Neurology in the Faculty of Health Science of the Medical University of Warsaw in Warsaw, Poland. The clinical diagnosis of PD was evaluated using the UK Parkinson’s Disease Society Brain Bank clinical diagnostic criteria at the time of specimen collection by two neurologists (DK and LM). A blood sample and a single 3 mm skin punch biopsy were collected.
4.2. Exome Sequencing in Sib-Pairs
Whole-exome sequencing (SureSelect Human All Exon v6 enrichment, Illumina NovaSeq 6000 platform, annotations according to Department of Medical Genetics, Institute of Mother and Child pipeline, VEP2.7) was performed on both affected family members [31]. Golden Helix SNP and Variation Suite (SVS) was used to annotate variants and identify shared variants between sib-pairs. Shared variants were filtered for coding variants excluding synonymous, potential splicing, and gnomAD European Non-Finnish minor allele frequency (MAF) < 0.01. The cosegregation of nominated variants was confirmed with Sanger sequencing in affected and nonaffected family members. The study design is summarized in Figure 2.
4.3. Replication Cohorts and Genotyping
Genotyping was performed in 5 different cohorts (2077 PD cases and 615 controls) collected from independent sites (Table 2). All individuals were of European Non-Finnish descent. The study was approved by the ethical review board from each institution and all participants provided informed written consent. A custom Applied Biosystems Taqman SNP Genotyping Assay (Thermo Fisher Scientific, Waltham, MA, USA) was designed for CTSB c.851G > T p.Gly284Val (NM_001908). Genotyping was performed using a QuantStudio 7 Real-Time PCR system. QuantStudio™ 7 Real-Time PCR Software (Thermo Fisher Scientific, Waltham, MA, USA) was used for analysis.
4.4. Gene Burden Analysis
The Mayo Clinic biobank control cohort consists of 885 unrelated samples of European Caucasian descent with no history of neurologic disease. The average age was 57 ± 15 (range 20–96) years with 438 (49.5%) males. The Mayo Clinic Florida familial PD cohort consists of 769 unrelated patients of European Caucasian descent diagnosed with PD and with a family history of PD [32]. The average age was 59 ± 18 (range 23–91) years with 462 (60%) males. All participants provided informed written consent prior to the commencement of this study. A gene burden analysis using SKAT was performed on rare (European, non-Finnish, MAF < 0.01) nonsynonymous variants with a CADD score greater than 20 within the CTSB gene (n = 16). Gender and age were used as covariates.
4.5. Protein Sequence and Structure Analysis
A multiple sequence alignment of human CTSB with a series of homologs from different species was performed in T-coffee Expresso [33,34]. The output was processed in ESPript 3 [35] to produce a structure-informed alignment. To visualize the location of Gly284, the crystal structure of human CTSB (PDB: 6AY2) [36], obtained via X-ray diffraction, was chosen for analysis in CCP4mg [37] (Figure 3).
4.6. Generation of Fibroblasts and Cell Culture
In an attempt to confirm the pathogenicity of CTSB p.Gly284Val, we compared the CTSB expression and activity using fibroblasts derived from patients and controls with matched age, APOE, and MAPT genotypes. Three independent experiments were performed for each functional analysis.
A piece of skin taken from the forearm under local anesthesia (size 1 mm × 1 mm) was cut and exposed to collagenase (24 h, 37 °C) (Sigma-Aldrich, St. Louis, MO, USA). After that, cells were seeded and cultured in Advance DMEM/F-12 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific), 1× Glutamax (Thermo Fisher Scientific), and 1× Antibiotic-Antimycotic agent (Thermo Fisher Scientific) at 37 °C in the incubator with 5.0% CO2. Cells were fed every 2–3 days with a fresh growth medium and split after reaching 90% confluence in proportion 1/4.
Primary human dermal fibroblasts collected from CTSB p.Gly284Val mutation carriers and controls (cryopreserved HDF cells; [Cell Applications Inc., San Diego, CA, USA, 106–05A]) were grown in Dulbecco’s modified Eagle medium (DMEM [Thermo Fisher Scientific, 11,965,118]) supplemented with 10% fetal bovine serum (FBS [Neuromics, FBS001800112]), 1% PenStrep (Thermo Fisher Scientific, 15,140,122) and 1% non-essential amino acids (Thermo Fisher Scientific, 11,140,050). All cells were grown at 37 °C, 5% CO2: air in the humidified atmosphere.
4.7. RNA Extraction and qRT-PCR
Total RNA isolation was performed using TRIzol® Reagent by Life Technologies (Carlsbad, CA, USA). Frozen tissue (55 mg) was homogenized in 1 mL of TRIzol reagent using a pestle and incubated for 5 min at room temperature. The 0.2 mL of chloroform was added, the sample was capped, mixed vigorously by hand for 15 s, and incubated at room temperature for 3 min. The samples were centrifuged at 12,000× g for 15 min at 4 °C. Following centrifugation, the aqueous phase was transferred to a fresh tube containing 0.5 mL of isopropyl alcohol and incubated at room temperature for 10 min followed by centrifugation at 12,000× g for 10 min at 4 °C. The pellet was washed once with 1 mL of 75% ethanol and centrifuged at 7500× g for minutes at 4 °C. The RNA pellet was left to air dry briefly and reconstituted using Nuclease-Free water. RNA quality was assessed using Agilent RNA 6000 Nano kit (Agilent Technologies, Waldbronn, Germany). The Hight-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Cheshire, UK) was used to convert total RNA to single-stranded cDNA. QuantStudio™ Real-Time PCR Software (Thermo Fisher Scientific, Waltham, MA, USA) was used for the gene expression analysis (Taqman® Gene Expression Assay, Probe ID Hs05518041_s1, FAM).
4.8. Protein Extraction and Western Blot
As previously described, cells were lysed in RIPA buffer containing protease inhibitor cocktail and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO, USA, 11697498001 and 04906837001) [38,39]. After 30 min incubation on ice, cell lysates were cleared for 15 min, 4 °C at 20,817× g, and protein concentrations were determined by BCA assay (Thermo Fisher Scientific, Waltham, MA, USA, 23225). Cell lysates containing 10 µg of protein were diluted in Laemmli buffer and boiled at 95 °C for 5 min before running on Tris-Glycine gels (Invitrogen, EC60485BOX). Post transfer of protein onto PVDF membranes (Millipore Sigma, St. Louis, MO, USA, IPVH00010), membranes were blocked in 5% skim milk (Genesee Scientific, El Cajon, CA, USA, 20–241) and incubated with primary antibodies against CTSB (Abcam, Cambridge, UK, 58802; 1:5000) and GAPDH (Meridian Life Sciences, Memphis, TN, USA, H86504M; 1:500,000) overnight at 4 °C. After washing, membranes were incubated in secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA, 715-035-150; 1:10,000) for 1 h at room temperature. Proteins were visualized using Immobilon Western Chemiluminescent HRP Substrate (Millipore Sigma, St. Louis, MO, USA, WBKLS0500) on Pro Signal Blotting film (Genesee Scientific, El Cajon, CA, USA, 30–810 L). Quantification of western blots was performed using Image Studio Lite software. The intensity levels of protein bands were background subtracted and then normalized to the loading control.
4.9. CTSB Enzymatic Activity Assay
The CTSB activity was compared between patient-derived fibroblasts and control fibroblasts using a commercially available CTSB activity kit assay (Abcam, Cambridge, UK, ab65300). The harvested cell pellets were resuspended in 50 µL of chilled Cell Lysis Buffer and incubated on ice for 30 min. Then, the samples were centrifuged at 4 °C for 4 min and the supernatants were collected. The protein concentration was determined by the BCA method. To measure the CTSB activity, cell lysates containing10 ug of proteins were loaded into a 96-well plate and incubated at 37 °C with 2 µL of 10 mM CB Substrate Ac-RR-AFC (Abcam, Cambridge, UK) and 50 µL of the activity assay buffer for 2 h. Fluorescence signals in the plate were measured by a 2104 EnVision® Multilabel Plate Reader (PerkinElmer, Inc., Waltham, MA, USA) at Ex/Em 400/505 nm.
4.10. Immunocytochemistry
As previously described, fibroblasts were seeded onto a PDL (Sigma-Aldrich, St. Louis, MO, USA, P0899) coated glass coverslips and fixed with 4% paraformaldehyde after attaching to the bottom [38,39,40]. Fibroblasts were immunostained with primary antibody against CTSB (Abcam, Cambridge, UK, 58802; 1:2000) followed by incubation with secondary antibody (Invitrogen, Waltham, MA, USA, A-11001; 1:1000) and Hoechst 33,342 (Invitrogen, Waltham, MA, USA, H21492l; 1:5000). Slices were imaged with an Axio-Observer microscope equipped with an ApoTome Imaging System (Zeiss, Oberkochen, Germany).
Acknowledgments
We thank all patients and family members. Mayo Clinic Florida is an APDA Center for Advanced Research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23137086/s1.
Author Contributions
L.M.M., X.H., J.M.B., F.C.F., L.T.C., A.I.S.-B., R.L.W., A.H.F., S.R.-N. and E.P. performed experiments and analyzed the data. A.J.S., M.B., M.B.-J., J.D., L.F., P.J., A.P.-C., K.K., A.K.-W., T.L., D.A.O., G.O., A.P., I.R., Y.S., J.S. (Joanna Siuda), M.W., J.S. (Jaroslaw Slawek), M.R.-B., R.U., A.H.F., M.F., S.S., P.J.M., D.K., O.A.R., D.H.-Z. and Z.K.W. contributed clinical and genetic information as well as materials of the patients. D.H.-Z., O.A.R., W.S. and Z.K.W. conceived and designed the study. L.M.M., X.H., W.S., A.I.S.-B. and O.A.R. wrote the manuscript. All authors contributed to drafting or revising the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was approved by the ethical review board from each institution.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data available on request due to restrictions eg privacy or ethical. The data presented in this study are available on request from the corresponding author. The data are not publicly available due to patient’s privacy.
Conflicts of Interest
There is no conflict of interest relevant to the study.
Funding Statement
L.M.M. is supported by the Polish National Agency for Academic Exchange Iwanowska’s Fellowship PPN/IWA/2018/1/00006/U/00001/01, the APDA, the Foundation for Polish Science (FNP) and the Haworth Family Professorship in Neurodegenerative Diseases Fund. X.H. is supported by pilot and developmental project grants from the Mayo Clinic Alzheimer Disease Research Center (ADRC) and a fellowship awarded by the American Parkinson Disease Association (APDA). F.C.F. is the recipient of fellowships from the Younkin Scholar Program and the APDA and is supported by the Michael J. Fox Foundation for Parkinson’s Research (MJFF), a Gerstner Family Career Development Award from the Mayo Clinic Center for Individualized Medicine, the Mayo Clinic Center for Biomedical Discovery (CBD), and the Florida Department of Health-Ed and Ethel Moore Alzheimer’s Disease Research Program [22A07]. A.H.F. is supported by a stipend from Mayo Clinic Graduate School of Biomedical Sciences. P.J.M. is partially supported by the National Institute of Neurological Disorders and Stroke (NIH/NINDS) [R01 NS085070, R01 NS110085 and U54 NS110435], the APDA Center for Advanced Research, the Mayo Clinic Foundation, and the American Brain Foundation. O.A.R. is partially supported by National Institute of Health/National Institute of Neurological Disorders and Stroke (NIH/NINDS) [R01 NS085070 and U54 NS110435], the Department of Defense Congressionally Directed Medical Research Programs (CDMRP) [W81XWH-17-1-0249], James Ester King Foundation FL state, the Michael J. Fox Foundation for Parkinson’s Research (MJFF), the American Parkinson Disease Association (APDA), and American Brain Foundation. W.S. is partially supported by the National Institute on Aging (NIH/NIA) [R56 AG062556], National Institute of Neurological Disorders and Stroke (NIH/NINDS) [R01 NS085070, R01 NS110085 and U54 NS110435], the Department of Defense Congressionally Directed Medical Research Programs (CDMRP) [W81XWH-17-1-0248], the Florida Department of Health-Ed and Ethel Moore Alzheimer’s Disease Research Program [9AZ10], the Michael J. Fox Foundation for Parkinson’s Research (MJFF), Mayo Clinic Foundation, the Mayo Clinic Center for Biomedical Discovery and the Robert and Arlene Kogod Center on Aging. Z.K.W. is partially supported by the NIH/NIA and NIH/NINDS (1U19AG063911, FAIN: U19AG063911), Mayo Clinic Center for Regenerative Medicine, Mayo Clinic in Florida Focused Research Team Program, gifts from the Sol Goldman Charitable Trust and Donald G. and Jodi P. Heeringa Family, the Haworth Family Professorship in Neurodegenerative Diseases fund, and the Albertson Parkinson’s Research Foundation. He serves as PI or Co-PI on Biohaven Pharmaceuticals, Inc. (BHV4157-206 and BHV3241-301), Neuraly, Inc. (NLY01-PD-1), and Vigil Neuroscience, Inc. (VGL101-01.001) clinical studies. He serves as an external advisory board member for Vigil Neuroscience. The study is partially supported by research grant from the National Science Center in Poland [grant number: 2017/01/X/NZ4/01450].
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Balestrino R., Schapira A. Parkinson disease. Eur. J. Neurol. 2020;27:27–42. doi: 10.1111/ene.14108. [DOI] [PubMed] [Google Scholar]
- 2.Postuma R.B., Berg D., Stern M., Poewe W., Olanow C.W., Oertel W., Obeso J., Marek K., Litvan I., Lang A.E. MDS clinical diagnostic criteria for Parkinson’s disease. Mov. Disord. 2015;30:1591–1601. doi: 10.1002/mds.26424. [DOI] [PubMed] [Google Scholar]
- 3.Dickson D.W. Neuropathology of Parkinson disease. Parkinsonism Relat. Disord. 2018;46:S30–S33. doi: 10.1016/j.parkreldis.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karimi-Moghadam A., Charsouei S., Bell B., Jabalameli M.R. Parkinson disease from mendelian forms to genetic susceptibility: New molecular insights into the neurodegeneration process. Cell. Mol. Neurobiol. 2018;38:1153–1178. doi: 10.1007/s10571-018-0587-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milanowski Ł.M., Ross O.A., Friedman A., Hoffman-Zacharska D., Gorka-Skoczylas P., Jurek M., Koziorowski D., Wszolek Z.K. Genetics of Parkinson’s disease in the Polish population. Neurol. Neurochir. Pol. 2021;55:241–252. doi: 10.5603/PJNNS.a2021.0013. [DOI] [PubMed] [Google Scholar]
- 7.Siuda J., Boczarska-Jedynak M., Budrewicz S., Figura M., Fiszer U., Gajos A., Gorzkowska A., Koziorowska-Gawron E., Koziorowski D., Krygowska-Wajs A., et al. Validation of the Polish version of the Movement Disorder Society-Unified Parkinson’s Disease Rating Scale (MDS-UPDRS) Neurol. I Neurochir. Pol. 2020;54:416–425. doi: 10.5603/PJNNS.a2020.0049. [DOI] [PubMed] [Google Scholar]
- 8.Wissemann W.T., Hill-Burns E.M., Zabetian C.P., Factor S.A., Patsopoulos N., Hoglund B., Holcomb C., Donahue R.J., Thomson G., Erlich H., et al. Association of Parkinson disease with structural and regulatory variants in the HLA region. Am. J. Hum. Genet. 2013;93:984–993. doi: 10.1016/j.ajhg.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porrini V., Mota M., Parrella E., Bellucci A., Benarese M., Faggi L., Tonin P., Spano P.F., Pizzi M. Mild Inflammatory Profile without Gliosis in the c-Rel Deficient Mouse Modeling a Late-Onset Parkinsonism. Front. Aging Neurosci. 2017;9:229. doi: 10.3389/fnagi.2017.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung S.J., Armasu S.M., Biernacka J.M., Lesnick T.G., Rider D.N., Cunningham J.M., Maraganore D.M. Variants in estrogen-related genes and risk of Parkinson’s disease. Mov. Disord. 2011;26:1234–1242. doi: 10.1002/mds.23604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan L., Song Z., Deng X., Zheng W., Guo Y., Yang Z., Deng H. Systematic analysis of genetic variants in Han Chinese patients with sporadic Parkinson’s disease. Sci. Rep. 2016;6:33850. doi: 10.1038/srep33850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Germer E.L., Imhoff S., Vilariño-Güell C., Kasten M., Seibler P., Brüggemann N., Klein C., Trinh J. The Role of Rare Coding Variants in Parkinson’s Disease GWAS Loci. Front. Neurol. 2019;10:1284. doi: 10.3389/fneur.2019.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGlinchey R.P., Lee J.C. Cysteine cathepsins are essential in lysosomal degradation of α-synuclein. Proc. Natl. Acad. Sci. USA. 2015;112:9322–9327. doi: 10.1073/pnas.1500937112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein C.L., Rovelli G., Springer W., Schall C., Gasser T., Kahle P.J. Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. J. Neurochem. 2009;111:703–715. doi: 10.1111/j.1471-4159.2009.06358.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee S.H., Lee S., Du J., Jain K., Ding M., Kadado A.J., Atteya G., Jaji Z., Tyagi T., Kim W.H., et al. Mitochondrial MsrB2 serves as a switch and transducer for mitophagy. EMBO Mol. Med. 2019;11:e10409. doi: 10.15252/emmm.201910409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Santamarina E., García-Ruiz P., Martínez-Rubio D., Ezquerra M., Pla-Navarro I., Puente J., Martí M.J., Palau F., Hoenicka J. Regulatory rare variants of the dopaminergic gene ANKK1 as potential risk factors for Parkinson’s disease. Sci. Rep. 2021;11:9879. doi: 10.1038/s41598-021-89300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedashiro S., Pastuhov S.I., Nishioka T., Watanabe T., Kaibuchi K., Matsumoto K., Hanafusa H. LRRK1-phosphorylated CLIP-170 regulates EGFR trafficking by recruiting p150Glued to microtubule plus ends. J. Cell. Sci. 2015;128:385–396. doi: 10.1242/jcs.169102. [DOI] [PubMed] [Google Scholar]
- 18.Greenbaum L., Rigbi A., Lipshtat N., Cilia R., Tesei S., Asselta R., Djaldetti R., Goldwurm S., Lerer B. Association of nicotine dependence susceptibility gene, CHRNA5, with Parkinson’s disease age at onset: Gene and smoking status interaction. Parkinsonism Relat. Disord. 2013;19:72–76. doi: 10.1016/j.parkreldis.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Dong W., Qiu C., Gong D., Jiang X., Liu W., Liu W., Zhang L., Zhang W. Proteomics and bioinformatics approaches for the identification of plasma biomarkers to detect Parkinson’s disease. Exp. Med. 2019;18:2833–2842. doi: 10.3892/etm.2019.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansen I.E., Ye H., Heetveld S., Lechler M.C., Michels H., Seinstra R.I., Lubbe S.J., Drouet V., Lesage S., Majounie E., et al. Discovery and functional prioritization of Parkinson’s disease candidate genes from large-scale whole exome sequencing. Genome Biol. 2017;18:22. doi: 10.1186/s13059-017-1147-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrow S.L., Schierding W., Gokuladhas S., Golovina E., Fadason T., Cooper A.A., O’Sullivan J.M. Establishing gene regulatory networks from Parkinson’s disease risk loci. Brain. 2022:awac022. doi: 10.1093/brain/awac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blauwendraat C., Reed X., Krohn L., Heilbron K., Bandres-Ciga S., Tan M., Gibbs J.R., Hernandez D.G., Kumaran R., Langston R. Genetic modifiers of risk and age at onset in GBA associated Parkinson’s disease and Lewy body dementia. Brain. 2020;143:234–248. doi: 10.1093/brain/awz350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGlinchey R.P., Lacy S.M., Huffer K.E., Tayebi N., Sidransky E., Lee J.C. C-terminal α-synuclein truncations are linked to cysteine cathepsin activity in Parkinson’s disease. J. Biol. Chem. 2019;294:9973–9984. doi: 10.1074/jbc.RA119.008930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ritonja A., Popovic T., Turk V., Wiedenmann K., Machleidt W. Amino acid sequence of human liver cathepsin B. FEBS Lett. 1985;181:169–172. doi: 10.1016/0014-5793(85)81136-4. [DOI] [PubMed] [Google Scholar]
- 25.Perera R.M., Zoncu R. The lysosome as a regulatory hub. Annu. Rev. Cell Dev. Biol. 2016;32:223–253. doi: 10.1146/annurev-cellbio-111315-125125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasiljeva O., Reinheckel T., Peters C., Turk D., Turk V., Turk B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr. Pharm. Des. 2007;13:387–403. doi: 10.2174/138161207780162962. [DOI] [PubMed] [Google Scholar]
- 27.Cao M., Luo X., Wu K., He X. Targeting lysosomes in human disease: From basic research to clinical applications. Signal Transduct. Target. Ther. 2021;6:1–28. doi: 10.1038/s41392-021-00778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cermak S., Kosicek M., Mladenovic-Djordjevic A., Smiljanic K., Kanazir S., Hecimovic S. Loss of cathepsin B and L leads to lysosomal dysfunction, NPC-like cholesterol sequestration and accumulation of the key Alzheimer’s proteins. PLoS ONE. 2016;11:e0167428. doi: 10.1371/journal.pone.0167428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang M., Meng J., Zeng F., Qing H., Hook G., Hook V., Wu Z., Ni J. Cathepsin B inhibition blocks neurite outgrowth in cultured neurons by regulating lysosomal trafficking and remodeling. J. Neurochem. 2020;155:300–312. doi: 10.1111/jnc.15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Man S.M., Kanneganti T.-D. Regulation of lysosomal dynamics and autophagy by CTSB/cathepsin B. Autophagy. 2016;12:2504–2505. doi: 10.1080/15548627.2016.1239679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milanowski Ł., Hoffman-Zacharska D., Geremek M., Friedman A., Figura M., Koziorowski D. The matter of significance–Has the p.(Glu121Lys) variant of TOR1A gene a pathogenic role in dystonia or Parkinson disease? J. Clin. Neurosci. 2020;72:501–503. doi: 10.1016/j.jocn.2019.12.018. [DOI] [PubMed] [Google Scholar]
- 32.Konno T., Deutschländer A., Heckman M.G., Ossi M., Vargas E.R., Strongosky A.J., van Gerpen J.A., Uitti R.J., Ross O.A., Wszolek Z.K. Comparison of clinical features among Parkinson’s disease subtypes: A large retrospective study in a single center. J. Neurol. Sci. 2018;386:39–45. doi: 10.1016/j.jns.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Notredame C., Higgins D.G., Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 34.Armougom F., Moretti S., Poirot O., Audic S., Dumas P., Schaeli B., Keduas V., Notredame C. Expresso: Automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34:W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei B., Gunzner-Toste J., Yao H., Wang T., Wang J., Xu Z., Chen J., Wai J., Nonomiya J., Tsai S.P., et al. Discovery of Peptidomimetic Antibody-Drug Conjugate Linkers with Enhanced Protease Specificity. J. Med. Chem. 2018;61:989–1000. doi: 10.1021/acs.jmedchem.7b01430. [DOI] [PubMed] [Google Scholar]
- 37.McNicholas S., Potterton E., Wilson K.S., Noble M.E. Presenting your structures: The CCP4mg molecular-graphics software. Acta Cryst. D Biol. Cryst. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puschmann A., Fiesel F.C., Caulfield T.R., Hudec R., Ando M., Truban D., Hou X., Ogaki K., Heckman M.G., James E.D., et al. Heterozygous PINK1 p.G411S increases risk of Parkinson’s disease via a dominant-negative mechanism. Brain. 2016;140:98–117. doi: 10.1093/brain/aww261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ando M., Fiesel F.C., Hudec R., Caulfield T.R., Ogaki K., Górka-Skoczylas P., Koziorowski D., Friedman A., Chen L., Dawson V.L., et al. The PINK1 p.I368N mutation affects protein stability and ubiquitin kinase activity. Mol. Neurodegener. 2017;12:32. doi: 10.1186/s13024-017-0174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watzlawik J.O., Hou X., Fricova D., Ramnarine C., Barodia S.K., Gendron T.F., Heckman M.G., DeTure M., Siuda J., Wszolek Z.K., et al. Sensitive ELISA-based detection method for the mitophagy marker p-S65-Ub in human cells, autopsy brain, and blood samples. Autophagy. 2021;17:2613–2628. doi: 10.1080/15548627.2020.1834712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on request due to restrictions eg privacy or ethical. The data presented in this study are available on request from the corresponding author. The data are not publicly available due to patient’s privacy.