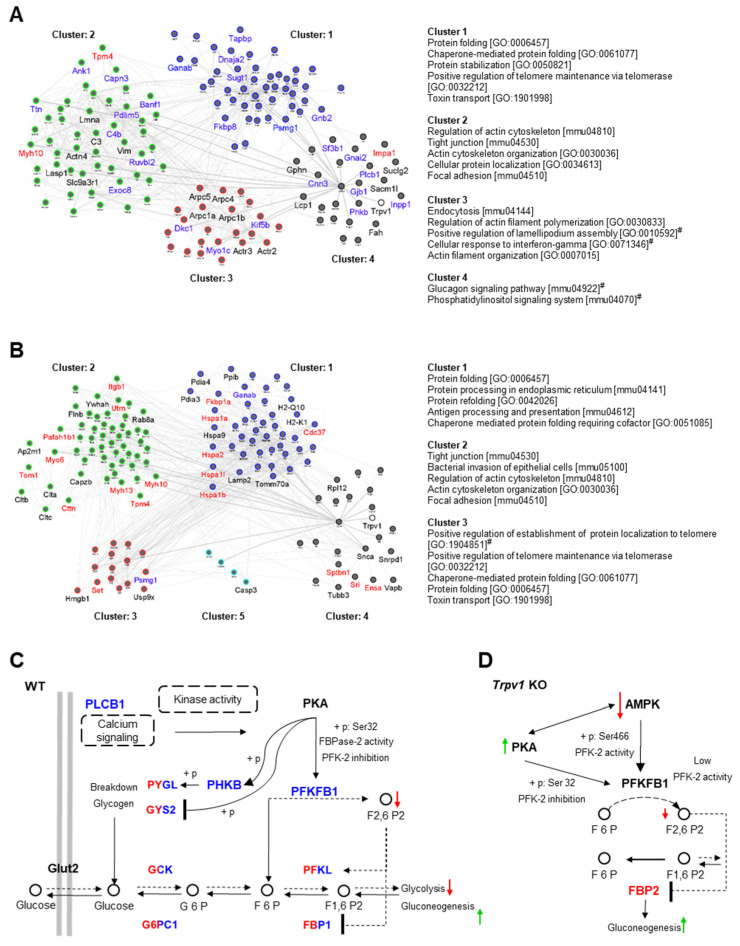
Figure 4.
Network propagation highlights important liver clustering pathways and biological networks in the absence of TRPV1 channels. Network propagation of the TRPV1 channel in the liver of WT (A) and Trpv1 KO mice (B). TRPV1 protein input under each network served as a driver for network propagation in Trpv1 KO and WT. Separately propagated subnetworks with 141 proteins found to be significant in the propagation were clustered into smaller subnetworks and annotated using integrated enrichment analysis (GO-BP and KEGG pathway) to identify functional networks that are different between two propagated subnetworks (EnrichmentMap FDR < 0.01). Node names represent the proteins exclusively found in each propagated subnetwork; black for proteins shared between Trpv1 KO and WT proteomes; blue for proteins only found or upregulated in the WT proteome; red for proteins only found or upregulated in the Trpv1 KO proteome. Cluster title indicates the top five most significant biological function(s) for each cluster, and (#) refers to the KEGG pathway/GO-BP annotations exclusively enriched in each propagated subnetwork. (C) Framework of propagated network integration for enriched glucagon signaling pathway from KEGG in the WT proteome. (D) Framework shows proposed modulation of FBP-2 by PKA and AMPK kinases in Trpv1 KO mice resulting in more active gluconeogenesis than in WT mice. Blue nodes for proteins only found in the WT proteome, dual colors (blue/red) for proteins shared among proteomes, and black for proteins not found in the proteomic data. (+p) refers to phosphorylation and (−p) to dephosphorylation.